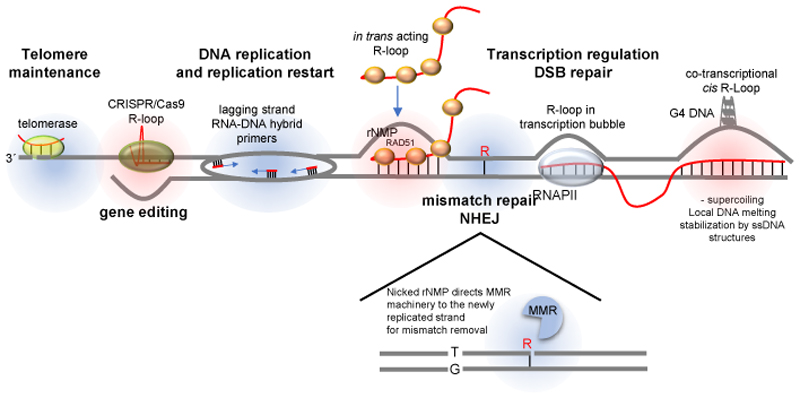
Figure 1. Types of RNA–DNA hybrids.
RNA has the capacity to localize to genomic regions in a sequence specific manner and regulate downstream cellular processes (labelled in bold). R-loops are three-stranded structures harbouring an RNA–DNA hybrid and a displaced strand of DNA (highlighted in red). Non-R-loop forming RNA–DNA hybrids (highlighted in blue) are involved in different chromosomal transactions. At telomeres, the RNA moiety of the telomerase holoenzyme base-pairs with the 3´overhang at chromosome ends and provides a template for their extension. DNA replication, especially on the lagging strand, is dependent on the prior synthesis of small RNAs, which are used as polymerization primers by DNA polymerases. The cleavage of DNA-incorporated ribonucleoside monophosphates (rNMPs) on the 5´side serves as a signal for the DNA mismatch repair (MMR) machinery to distinguish the newly replicated strand from the template strand and ensure that the right mismatched base (here, ‘T’) is removed. R-loops are thought to form behind the RNA polymerase transcription machinery, where negative DNA supercoiling results in DNA unwinding, which provides an opportunity for the 5´end of the nascent transcript to base-pair with the template strand. Hybrid formation maybe be facilitated in the presence of G-rich sequences on the non-template strand, which may result in G4 structures. When RNA is produced from an exogenous source (such as a plasmid) or from a homologous chromosome or a distal homologous sequence, R-loops can also form in trans in a RAD51 recombinase-dependent manner. All RNA molecules are depicted in red, and DNA is depicted in blue. DSB, double strand break; G4, G-quadruplex secondary structure; NHEJ, non-homologous end joining; Pol II, RNA polymerase II.