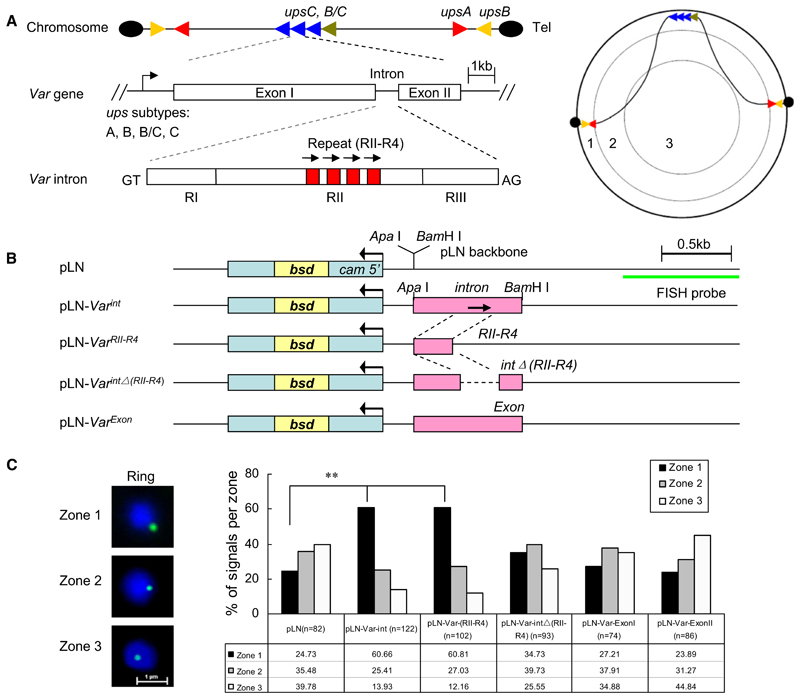
Figure 1. Var Introns Target Episomes to the Nuclear Periphery.
(A) Genomic organization, structure, and nuclear position of var genes. (Left panel) upsB or upsA corresponds to subtelomeric and upsC corresponds to chromosome internal var gene members. Each subtype var gene is labeled with a different color. Var introns contain up to four 18 bp repetitions in its region II (RII) domain. (Right panel) Schematic illustration of subnuclear localization of various subtype var genes on different chromosome sites. The area of the nuclear section was divided into three concentric zones with equal surfaces, and nuclear periphery corresponds to the zone 1.
(B) pLN vector constructs with 3D7 upsC-type full-length intron (PFD1000c), the intron repeat region (RII-R4), the intron without the RII-R4 region, two distinct regions from var exon I and exon II, and the vector backbone control. The drug-selectable marker gene (bsd) and the region used as FISH probe are shown, respectively.
(C) FISH signals of episomes (green) were localized into zones 1, 2, or 3 with equal surface (see A, right panel). The bar graph represents the position of FISH signals with respect to three concentric zones. DAPI (blue), nuclear DNA. n, number of nuclei analyzed from two independent ring stage samples. **p < 0.001 (χ2 test).