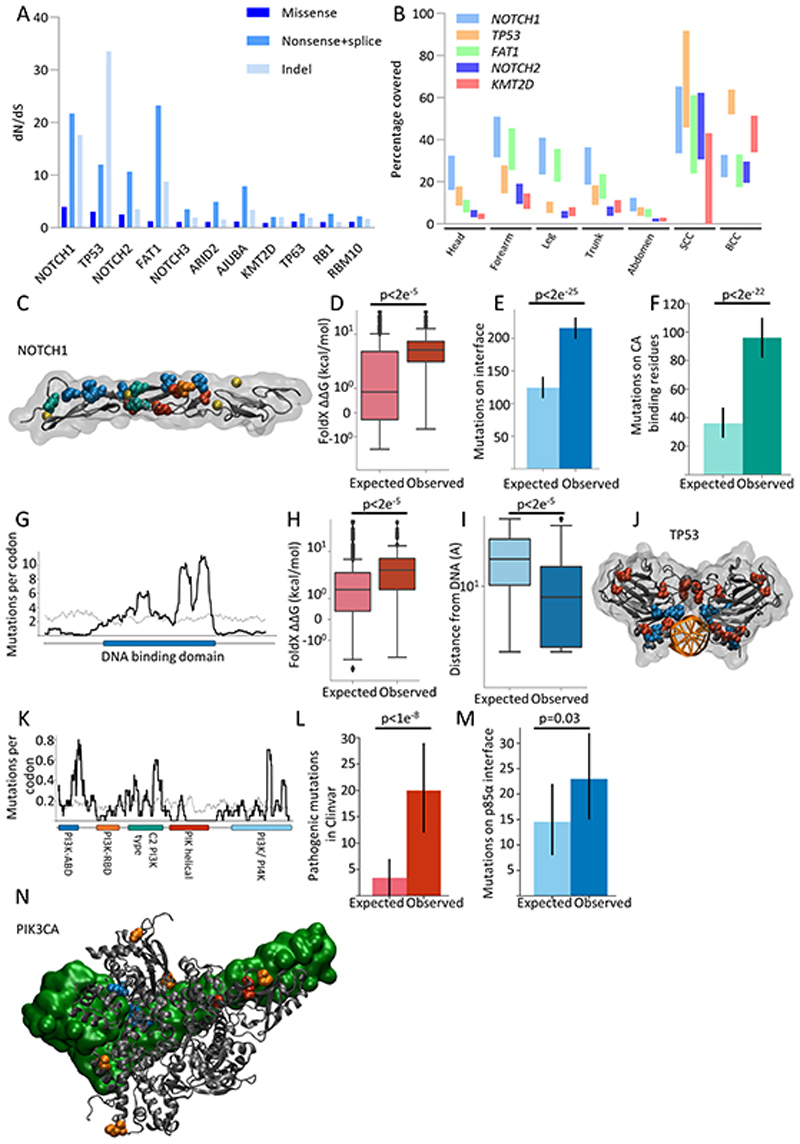
Figure 2. Positive selection of genes linked with cancer in normal skin.
a. dN/dS ratios for missense, nonsense/splice substitutions and insertions/deletions (indel) across all body sites for genes under significant (global q <0.01 and dN/dS>2) positive selection.
b. Estimated percentage of cells carrying mutations in the most strongly positively selected genes (NOTCH1, FAT1, TP53 and NOTCH2) for each body site as well as for basal and squamous cell carcinomas (see methods). Upper and lower bound range allows for uncertainty in copy number and biallelic mutations. Upper bound represents no CNA and one mutant allele per gene.
c-f: Positive selection of categories of missense mutations in NOTCH1 EGF repeats 11-12 that form part of the ligand binding domain:
c. Structure of NOTCH1 EGF11-13 (PDB 2VJ3). Residues containing missense mutations that occur >10 times are highlighted. Ligand binding interface residues, blue; calcium binding residues, green; destabilising residues, red; D464N, orange, does not fit into the previous categories. Calcium ions shown in yellow.
d. Missense mutations that are not on ligand-interface or calcium binding residues are significantly more destabilising than would be expected under neutral selection (p<2e−5, n=452, two-tailed Monte Carlo test, methods).
e. Non-calcium binding missense mutations with ΔΔG < 2kcal/mol (i.e. are not highly destabilising) occur on the ligand-binding interface significantly more than would be expected under neutral selection (p=2e−25, n=315, two-tailed binomial test, error bars show 95% confidence intervals, methods).
f. Missense mutations with ΔΔG < 2kcal/mol (i.e. are not highly destabilising) and that are not on the ligand-binding interface occur on calcium binding residues significantly more than would be expected under neutral selection (p=2e−22, n=195, two-tailed binomial test, error bars show 95% confidence intervals, methods).
g-h: Positive selection of missense mutations in TP53
g. Sliding window plot of missense mutations per codon in TP53. Observed counts shown by the black line. Expected counts assuming that missense mutations were distributed across the gene according to the mutational spectrum (methods) shown in grey. DNA-binding domain (DBD) of TP53 shown in blue below the x-axis.
h. Missense mutations in the TP53 DBD that are more than 5Å from the DNA are significantly more destabilising than would be expected under neutral selection (p<2e−5, n=760, two-tailed Monte Carlo test, methods).
i. Missense mutations with ΔΔG < 2kcal/mol (not highly destabilising) in the TP53 DBD are significantly closer to the DNA than would be expected under neutral selection (p<2e−5, n=395, two-tailed Monte Carlo test, methods).
j. Structure of the TP53 DNA-binding domain (PDB 2AC0) bound to DNA (orange). Residues containing missense mutations that occur at least 10 times are highlighted. Highly destabilising mutations (ΔΔG >= 2 kcal/mol) shown in red. Non-destabilising mutations shown in blue.
k-n: Positive selection of missense mutations in PIK3CA
k. Sliding window plot of missense mutations per codon in PIK3CA. Observed counts shown by the black line. Expected counts assuming that missense mutations were distributed across the gene according to the mutational spectrum (methods) shown in grey. Domains of PIK3CA encoded protein are shown below the x-axis.
l. Significantly more single nucleotide substitutions in PIK3CA are annotated as pathogenic/likely pathogenic in the Clinvar database than would be expected under neutral selection (q=1e−8, n=216, two-tailed binomial test, error bars show 95% confidence intervals, methods).
m. Significantly more missense mutations in PIK3CA occur in codons at the interface binding PIK3R1 (defined as PIK3CA residues with atoms within 5Å of PIK3R1 in PDB 4L1B) than would be expected under neutral selection (p=0.03, n=157, two-tailed binomial test, error bars show 95% confidence intervals, methods).
n. Structure of PIK3CA protein, grey, bound to PIK3R1, green (PDB 4L1B). Residues with mutations occurring at least 3 times are highlighted. Mutations close to PIK3R1 shown in blue, other mutations that are annotated as pathogenic/likely pathogenic shown in red, all others shown in orange.