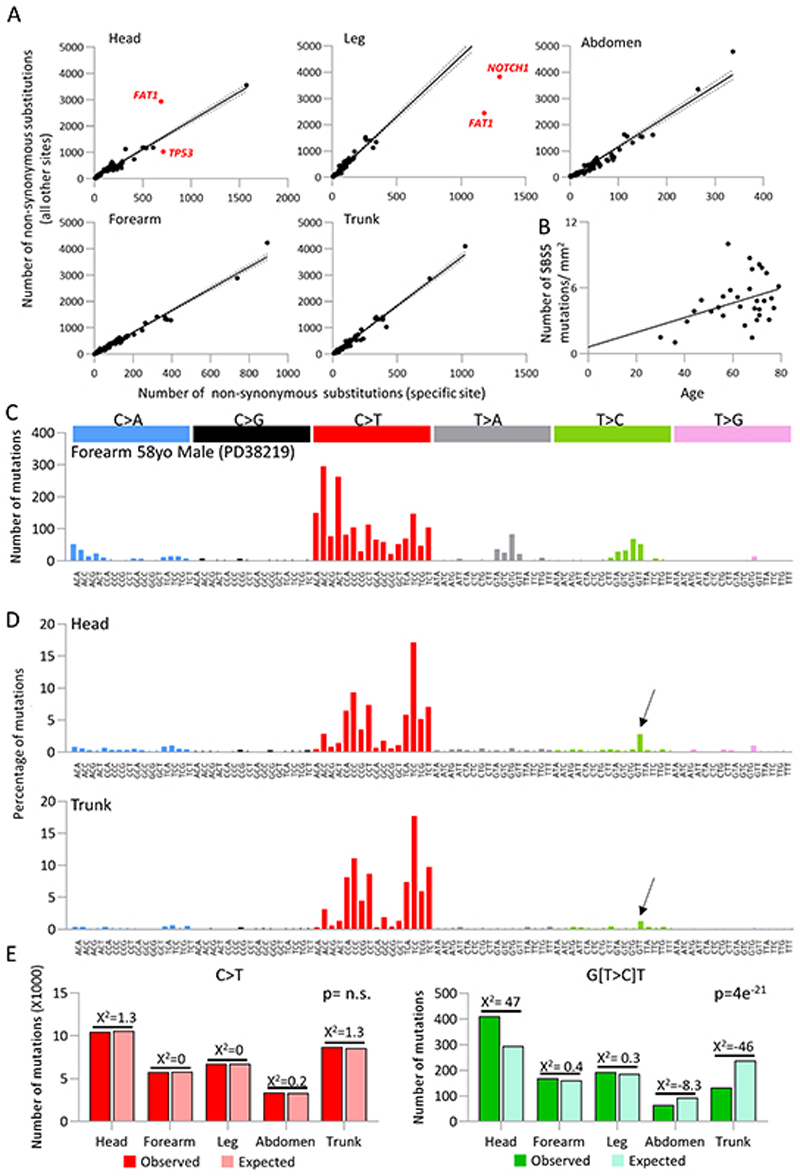
Figure 4. Human epidermis shows selection and signature variation between body sites.
a. Differential selection of TP53, NOTCH1 and FAT1 across different body sites. Number of non-synonymous mutations per gene from indicated body site versus all non-synonymous mutations in that gene from all other body sites. Each dot represents a gene from the 74 gene targeted bait panel. Solid line represents trendline with 95% CI marked (dotted line). For points highlighted in red, q<1×10−6 using a likelihood ratio test of dN/dS ratios (methods).
b. Correlation between patient age and number of mutations attributed to signature SBS5. Linear regression p=0.0299, slope 0.067, intercept 0.608
c. Trinucleotide spectrum for single base substitutions of donor PD38219 which shows considerable differences to all other donors. This signature is consistent with SBS32 (cosine similarity 0.95) linked with azathioprine treatment.
d. Combined trinucleotide spectra for single base substitutions in all samples from the head (top panel) and trunk (bottom panel) from targeted sequencing. Arrow indicates G(T>C)T peak which contributes to a higher proportion of overall burden in the head. Head = 14561 mutations, Trunk = 10515 mutations.
e. Chi-squared test for all C>T mutations versus G(T>C)T mutations by body site (chi-squared 105, df=4, p=0). A higher proportion of G(T>C)T mutations are observed at sites with higher relative risk of cSCC.