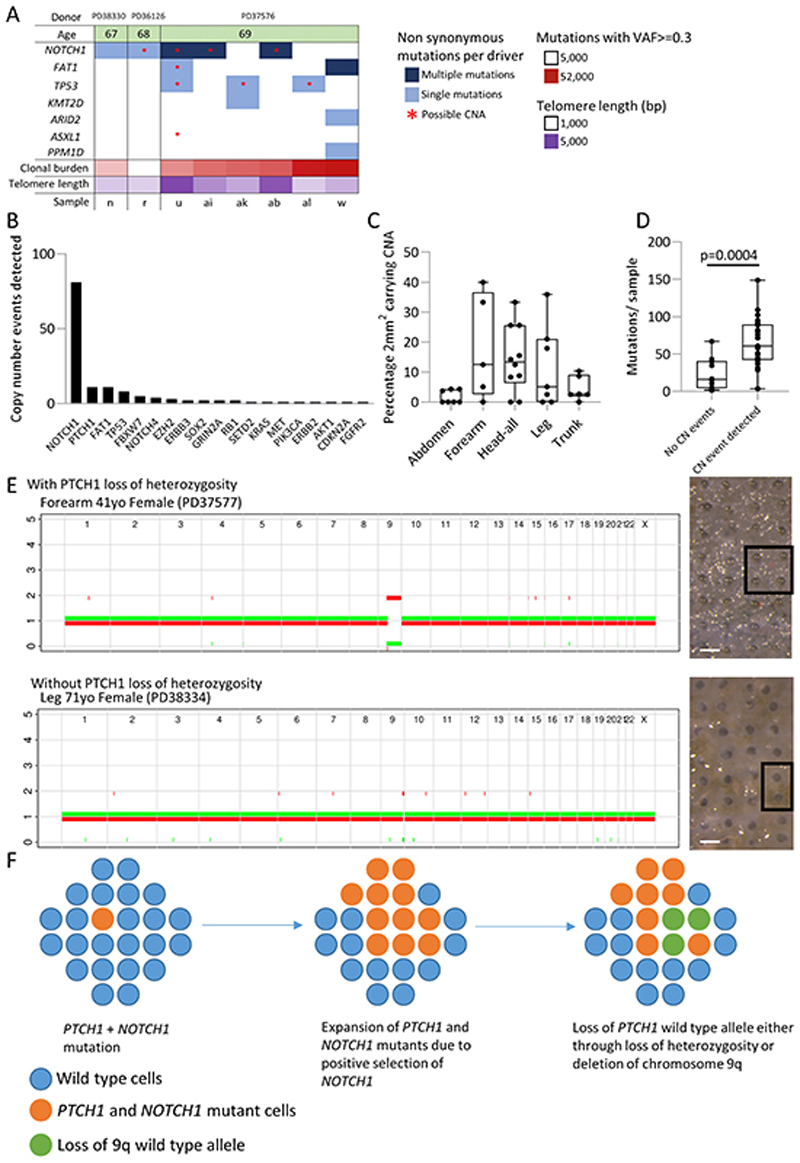
Figure 6. Normal human skin shows frequent copy number changes including loss of heterozygosity of PTCH1 .
a. Plot summarising the mutations (VAF>=0.3) and copy number alterations for genes identified as being under positive selection in targeted sequencing data for eight clonal whole-genome 2 mm2 grid samples. Age of donor, number of clonal (VAF>=0.3) mutations and telomere length for each sample is also shown. Not all events are independent since some samples are part of the same clone. Sample PD37576u shows multiple changes with loss of heterozygosity at 4q (FAT1), 9q (NOTCH1), 17p (TP53) and 20q (ASXL1).
b. Number of copy number events per gene detected by SNP phasing of targeted sequence data in all 1261 2mm2 grid samples.
c. Percentage of grids carrying a copy number event detectable by SNP phasing segregated by body site. Each dot represents an individual.
d. Average number of mutations per 2mm2 grid in patients either carrying or not carrying a copy number event p=3.8×10−4, Student’s two tailed t-test
e. Copy number profile of whole genome sequenced punch samples showing chromosome 9 loss of heterozygosity. The top example shows loss of heterozygosity for both NOTCH1 and PTCH1; the bottom example shows just NOTCH1 LOH. Scale bar = 0.5mm
f. Possible origin of basal cell carcinoma. In a wild type population of cells (blue circles) a single cell acquires PTCH1 and then NOTCH1 non-synonymous mutations (marked orange), the clone size expands and persists due to NOTCH1 positive selection. At a later time point the wild type PTCH1 allele is lost either through deletion or loss of heterozygosity (marked green), thus leaving a clone lacking functional PTCH1 expression and primed for BCC transformation.