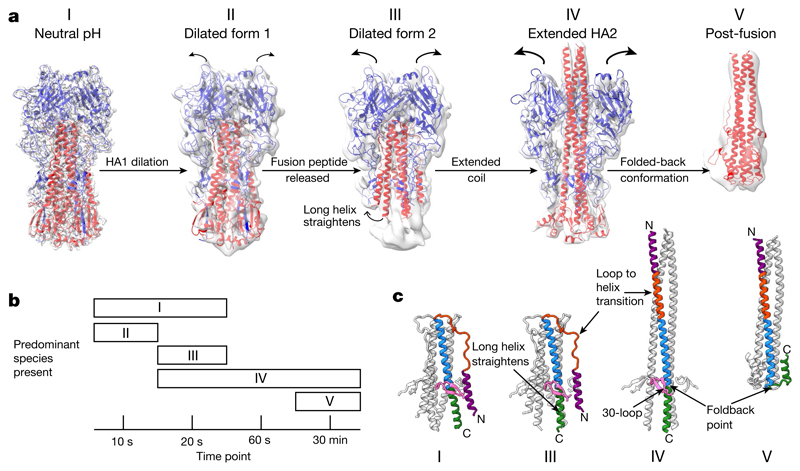
Fig. 1. HA fusion intermediates.
a, Cryo-EM reconstructions of HA at fusion pH from various time points after acidification. Cryo-EM maps are shown (grey) with models of HA1 (blue) and HA2 (red). Structures for states I (indistinguishable from the neutral-pH state), III and IV are from 20 s, and for state II from 10 s. State V was obtained with a 30-min incubation, supplemented with 2-mercaptoethanol, dissociating the disulfide-linked HA1. The state V model shown is the previously determined crystal structure of fusion-pH HA2 (Protein Data Bank (PDB, https://www.ebi.ac.uk/pdbe/) code 1HTM; ref. 7). b, Distribution of different species at chosen time points. c, Rearrangements in HA2 residues 38–125 associated with conformational states I, III, IV and V. The 30-loop (HA1 residues 22–37) is in pink; HA2 is coloured by residue number: purple, 38–55; orange, 56–75; blue, 76–105; green, 106–125.