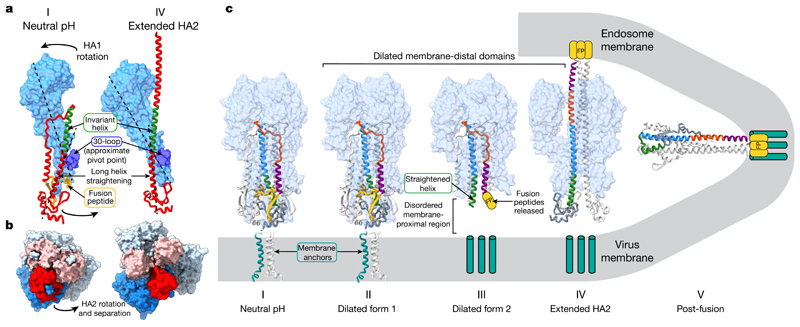
Fig. 3. Structural rearrangements of HA fusion intermediates.
a, Concerted rearrangements of HA1 and HA2 between states I and IV. HA1 is shown as a molecular surface in light blue, with the 30-loop in dark blue. HA2 is shown as a red ribbon with the invariant helix in green. The fusion peptide in state I is coloured yellow, but not ordered in state IV. Dotted lines show the trajectory of the membrane-proximal region of the extended helix in one case and the approximate long axis of HA1 in the other. HA1 rotates as a rigid body as the long helix of HA2 straightens relative to state I. This concerted motion is transmitted by the attachment of HA1 to HA2 via the 30-loop, which is the approximate pivot point of the HA1 rotation. The HA2 helix straightens into the space previously occupied by the now-displaced fusion peptide. b, Orthogonal view to a; membrane-proximal regions of HA2 open upon this concerted motion, transitioning from a closely packed neutral-pH conformation (state I) to the opened extended HA2 form (state IV). Images are molecular surfaces, with the monomers coloured as in a. c, Diagram showing HA membrane-fusion intermediates. HA1 is shown as a molecular surface with HA2 as a ribbon. One HA2 monomer is coloured by sequence: yellow, residues 1–37; purple, 38–55; orange, 56–75; blue, 76–105; green, 106–125; grey, 125–175. The location of the fusion peptide, not resolved in states III–V, is shown schematically (yellow). Membrane anchors (cyan), not present in the protein used here, are represented as blocks in the virus and endosome membranes in states III and IV. For states I and II the membrane anchor is that of another HA subtype, H1 (ref. 22). State IV is depicted as interacting with the virus membrane (via membrane anchors) and the endosomal membrane (via the fusion peptide) before refolding to state V, where the fusion peptide and membrane anchors are shown as colocated.