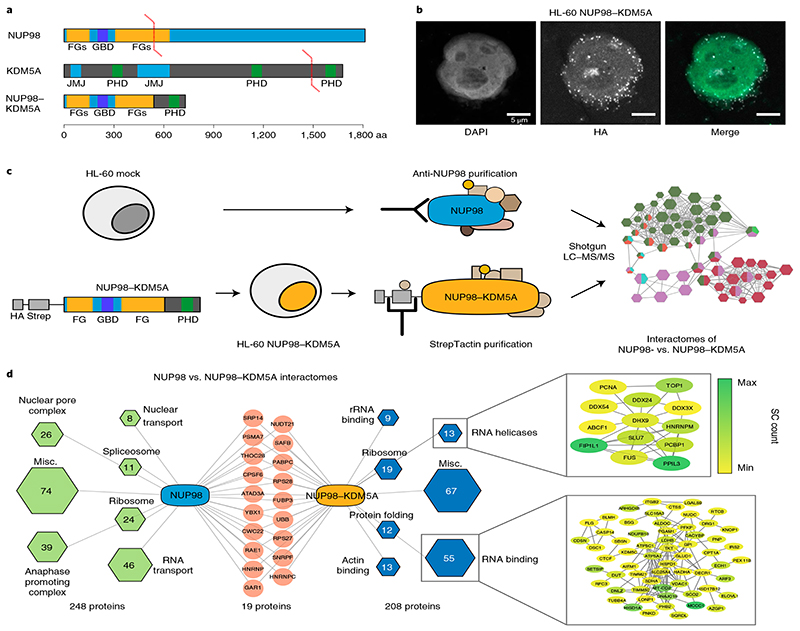
Fig. 1. NUP98–KDM5A does not operate in the context of the nuclear pore complex.
a, Domain architecture of endogenous NUP98 and KDM5A proteins and the oncogenic NUP98–KDM5A fusion protein. GBD, Gle2-binding domain; JMJ, Jumonji domain. b, Confocal microscopy images showing a representative HL-60 cell expressing NUP98–KDM5A, stained with DAPI (green in the merged image) and anti-HA antibody for fusion proteins (white in the merged image). Scale bar, 5μm. Six independent experiments were performed with similar results. c, Schematic representation of the experimental setup. HL-60 cells were transduced with a retroviral vector expressing tagged NUP98–KDM5A (left). Endogenous NUP98 complexes were pulled down from mock-transduced HL-60 cells with a NUP98-specific antibody, whereas tagged NUP98–KDM5A complexes were purified using Strep-Tactin (middle). Data on purified protein complexes were acquired by LC–MS/MS and subsequently analyzed (right). d, Interactome analysis was performed by CRaPome (antibody IP) or mock pull-down (Strep-Tactin AP) subtraction for NUP98 and NUP98–KDM5A, respectively. Individual protein complexes within the interactomes were obtained by K-means clustering (K=7) based on String db interactions, assigned using Gene Ontology Biological Processes and illustrated as hexagons (green for NUP98 and blue for NUP98–KDM5A complexes). Hexagon sizes and numbers represent the identified proteins associated with respective subcomplexes. Red nodes show shared proteins between the interactomes. Detailed string db networks (cutoff 0.4) are shown for subcomplexes of ‘RNA helicases’ and ‘RNA binding,’ and individual proteins are highlighted (yellow to green) according to abundance in MS acquisition.