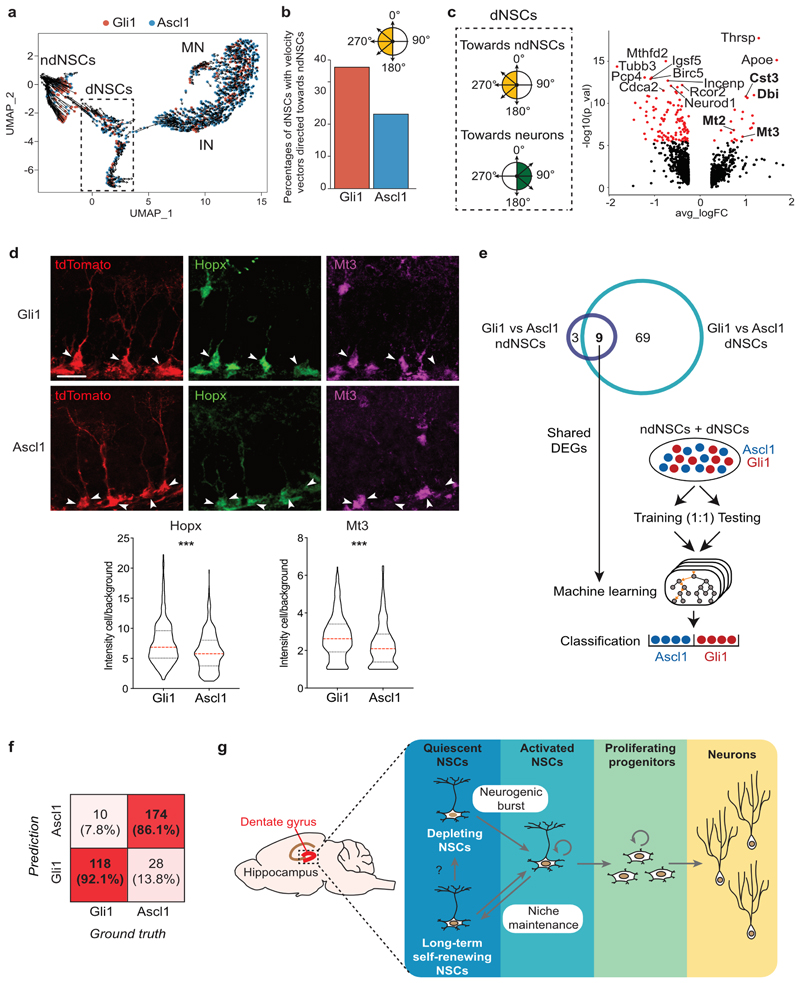
Fig. 4. Molecular profiling reveals distinct features of Gli1- vs. Ascl1-targeted NSCs.
a, The observed and the extrapolated future states (arrows) of Gli1 (red) or Ascl1 (blue) cells was calculated using RNA velocity. The velocities are visualized on the UMAP plot from panel (Fig. 3b). Velocity estimates based on nearest-cell pooling (k=300) were used. b, Percentages of dNSCs Gli1 (red) or Ascl1 (blue) with a velocity vector directed towards ndNSCs (yellow quadrants, vector angles between 180° and 380°). c, Volcano plot showing significantly DEGs (red, padj < 0.05) between dNSCs displaying an RNA velocity vector directed towards ndNSCs (yellow quadrants, vector angles between 180° and 380°), or towards IN (green quadrants, vector angles between >0° and <180°). Bolded gene names highlight common DEG with volcano plots (Fig. 3h) and (Fig. 3i). d, Top: representative examples of Gli1 and Ascl1 R cells stained using 4i protocol, 5 days post tamoxifen injection. Visualization of single channels: tdTom/red, Hopx/green, Mt3/magenta. Bottom: quantification of Hopx (Gli1 = 7.62 ± 0.197, n = 337 cells; Ascl1 = 6.14 ± 0.177, n = 316 cells; Unpaired t-test: t = 5.532; df = 651; ***p < 0.0001, two-tailed) and Mt3 (Gli1 = 2.72 ± 0.064, n = 337 cells; Ascl1 = 2.34 ± 0.065, n = 318 cells; Unpaired t-test: t = 4.201; df = 653; ***p < 0.0001, two-tailed) protein levels (fluorescence intensity of the cell/background) in Gli1- and Ascl1- targeted R cells. e, Schematic of machine learning approach. ndNSCs and dNSCs from Gli1- and Ascl1- targeted cells were divided into a training and a testing set (1:1). The 9 DEGs found in both ndNSCs (Fig. 3h) and dNSCs (Fig. 3i) comparisons were used to build three different machine learning models for classification: Random Forest Classifier (RFC), k Nearest Neighbor (k-NN) and Generalized Linear Models (GLM). The genotype (Gli1 or Ascl1) of ndNSCs and dNSCs was than predicted using the three classification models. f, Numbers and prediction accuracy rates to classify Gli1- vs. Ascl1-targeted cells using RFC, k-NN and GLM models together. Shades of red represent the accuracy of the prediction. g, Schematic showing the differential behavior of the two NSC populations in the DG: the first one gives rise to a burst of neurogenic activity followed by the depletion of the NSC. The second one is able to perform long-term self-renewal contributing to stem cell niche maintenance. Values are reported as mean ± s.e.m. Bars in violin plots represent median and quartiles. Scale bars represent 20 μm (d). For detailed statistics, see Supplementary Table 5.4