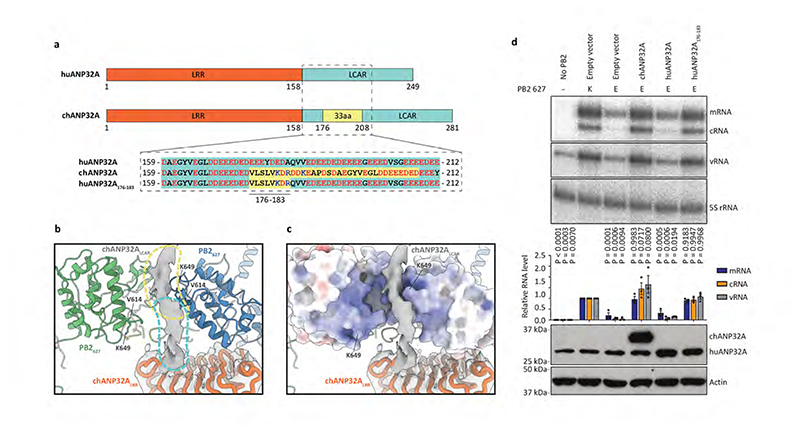
Fig. 3. Interaction of ANP32LCAR with FluPolC and the effect of ANP32A on FluPolA activity.
a, Schematic of huANP32A and chANP32A highlighting the 33 amino acid (33 aa) insertion in chANP32A and sequence differences. b, Close-up view of the cryo-EM density attributed to chANP32A (grey, threshold 0.934) with the chANP32ALRR domain represented in cartoon (orange) and the positions of PB2627 of FluPolR (blue) and FluPolE (green). Regions potentially corresponding to the N-terminal part of ANP32ALCAR and part of the avian 33 aa insertion are highlighted in cyan and yellow, respectively. V614 and K649 in FluPolC corresponds to K/Q591 and K/E627 in FluPolA, respectively. c, Same view as shown in (b), with the PB2627 of FluPolR and FluPolE shown in surface representation. d, Effect of chANP32A and huANP32A, wild type or mutant (huANP32A176-183), on the activity of FluPolA with mammalian-adapted PB2 K627 or avian-signature PB2 E627 in a viral RNP reconstitution assay in mammalian HEK 293T cells. Data of primer extension analysis of viral RNA levels with quantitation (upper panels) and western blot analysis of ANP32A levels (lower panel) with molecular weight markers is shown. Data are presented as mean values ± s.e.m. n = 3 biologically independent samples from n = 3 independent experiments. Ordinary one-way ANOVA with Dunnett’s post hoc test for multiple comparisons. P < 0.05 is considered significant to reject the null hypothesis. Western blot analyses were repeated from n = 3 independent experiments with similar results. For gel source data, see Supplementary Fig. 2.