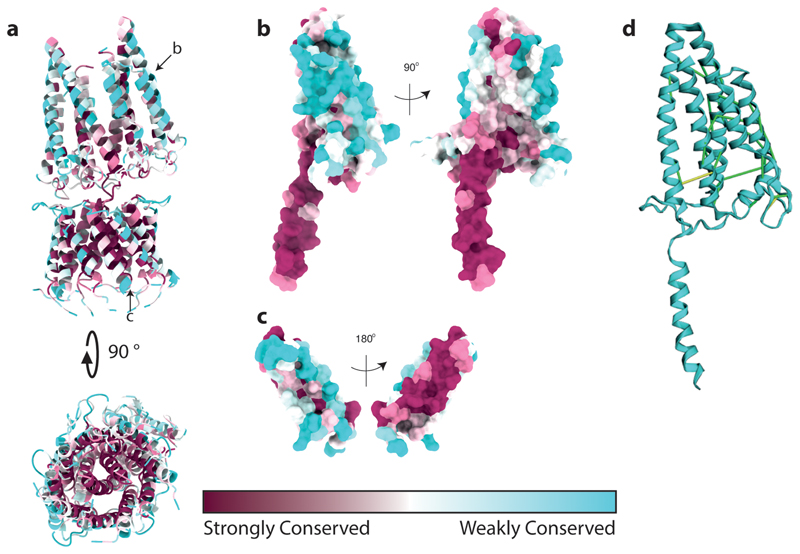
Extended Data Fig. 7. Conservation analysis of the GldLM′ complex.
a-c, Sequence conservation assessed using Consurf57.
a, The whole complex in cartoon representation. The chains shown individually in b and c are indicated with black arrows.
b,c, GldM (chain B) and GldL (chain F) in surface representation. The left hand panels show the chains in the same orientation as the upper panel of a.
d, Co-varying residue pairs in the first periplasmic domain of GldM identified using the Gremlin server64. The highest-scoring pairs (score cut-off 1.63) are shown on the structure of the GldLM′ complex. Contacts with a minimum atom-atom distance of <5 Å are shown in green, and 5-10 Å in yellow. Note, that no high-scoring GldM-GldM intermolecular pairs were observed for D1. Conversely dimerization of the other periplasmic domains is supported by co-variance with the following strongly co-varying pairs identified in the other domains D2:Val253-Ile284, Phe248-Glu-293, Tyr236-Arg261; D3: Thr327-Ala349, Asp331-Lys371; D4: Ala456-Asp483.