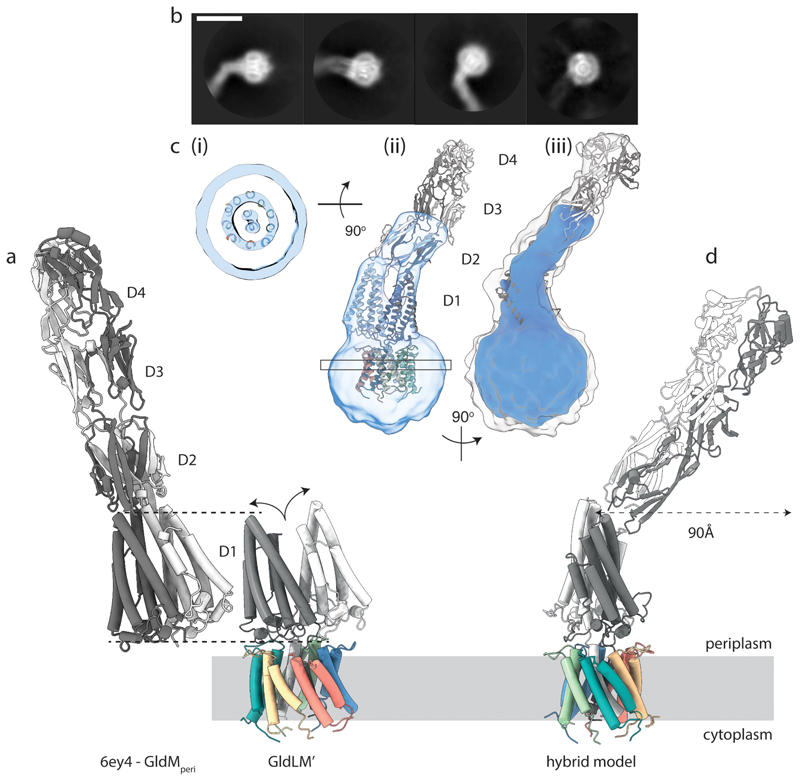
Fig. 3. Structure of the complete GldLM motor complex.
a, Comparison of the crystal structure of the periplasmic domains of GldM (PDB 6ey4, GldMperi) with the cryoEM structure of the periplasmically-truncated GldLM′ complex. The structures are displayed with the first periplasmic domain of the dark grey GldM chain aligned (dotted lines) and demonstrate the splaying (curved arrows) of the GldM chains in the context of the truncated complex. The GldL subunits are individually coloured as in Fig. 2a-c.
b, Representative 2D classes from micrographs of the full-length P. gingivalis motor (PorLM) showing detail in the transmembrane region (all 4 panels), a large bend between the first two periplasmic domains of PorM (left three panels), and splaying of the first periplasmic domains of the two copies of PorM (second panel). Scale bar 100 Å.
c, Different views of the 3D cryoEM volume of the P. gingivalis PorLM motor at a high (blue) and low (white in iii) contour level. (i) The PorM TMHs can be resolved allowing confident placement of the F. johnsoniae GldLM′ model in the volume. (ii) Splaying of the PorM first periplasmic domain pair is seen as in the F. johnsoniae GldLM′ complex. (iii) The extension to the volume seen at a low contour level matches well to the length and shape of the dimeric GldM periplasmic domains 2-4 seen in the crystal structure. See also Extended Data Fig. 8.
d, Composite model for the GldLM complex produced by combining the cryoEM structure of the GldLM′ complex with the crystallographic structures of the GldM periplasmic domains D2-D4. The bend between periplasmic domains 1 and 2 seen in the low resolution cryoEM volume would allow the tip of a rotating GldM dimer to define a large track at the outer membrane. The subunits are coloured as in panel a.