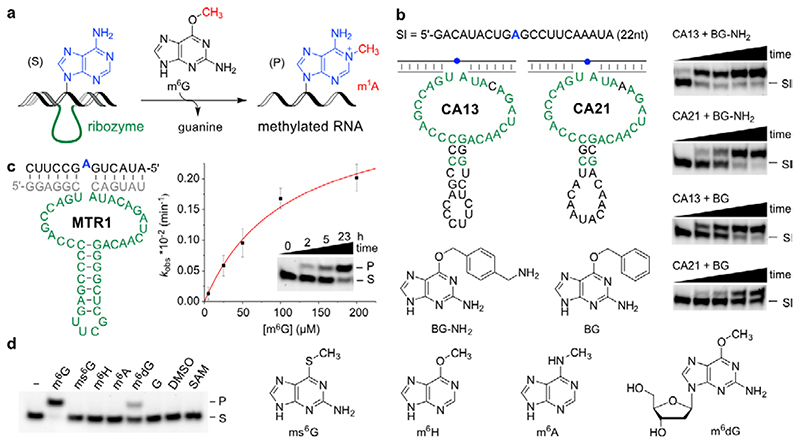
Fig. 1. Methyltransferase ribozyme-catalysed synthesis of m1A in RNA using m6G as methyl group donor.
a. Reaction scheme with intermolecular hybridization of ribozyme to target RNA. b. Sequences and predicted secondary structure of CA13 and CA21 ribozymes identified by in vitro selection, and their trans-activity for modification of a 22-nt RNA (Sl) with BG-NH2 or BG, analysed by 20% denaturing PAGE (100 µM guanine derivative, 40 mM MgCl2, pH 7.5, 37°C, timepoints 0, 0.5, 1, 2, 5 h). Representative images of three independent experiments with similar results. c. Methyltransferase ribozyme MTR1 with stabilized stem-loop shows efficient methyl group transfer. The insert shows a gel image of a 3'-fluorescein-labeled 13-mer RNA substrate (S) reacted with MTR1 and m6G (100 µM). k obs was determined with a 3'-fluorescein-labeled 17-mer RNA at five m6G concentrations ranging from 5–200 µM. The red line represents a curve fit to k obs = k max[m6G]/(k m,app+[m6G]). Individual data points (white, n = 3), mean ± s.e.m. (black). d. Structures of m6G analogues tested. Gel image shows that product formation only occurs with m6G, and to a minor extent with m6dG (24 h reaction time, 25°C, with 100 µM m6G or analog). Representative image from two independent experiments. DMSO = dimethyl sulfoxide, G = guanine, SAM = S-adenosylmethionine.