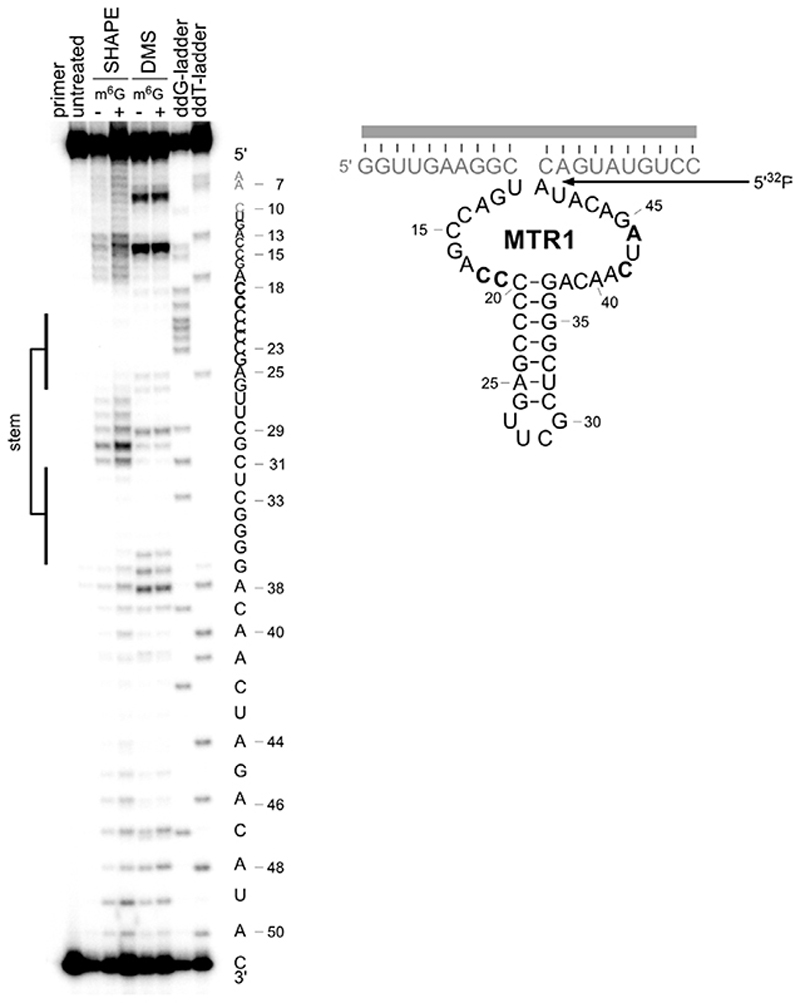
Figure 3. Extended Data Figure 3.
RNA structure probing by DMS and SHAPE. MTR1 (Rz3) was annealed with 17-nt RNA (R6), treated with dimethylsulfate (DMS) or 1-methyl-7-nitroisatoic anhydride (1M7), in presence (+) or absence (−) of m6G, and the modification pattern was analysed by primer extension (5'-32P-labeled D4) with Superscript III. DMS probes the accessibility of the Watson-Crick face of A and C, while SHAPE with 1M7 probes the flexibility of the backbone. Both probing methods confirm the central base-paired stem and reveal the protection of several additional nucleotides (bold). The experiment was repeated three times with similar results.