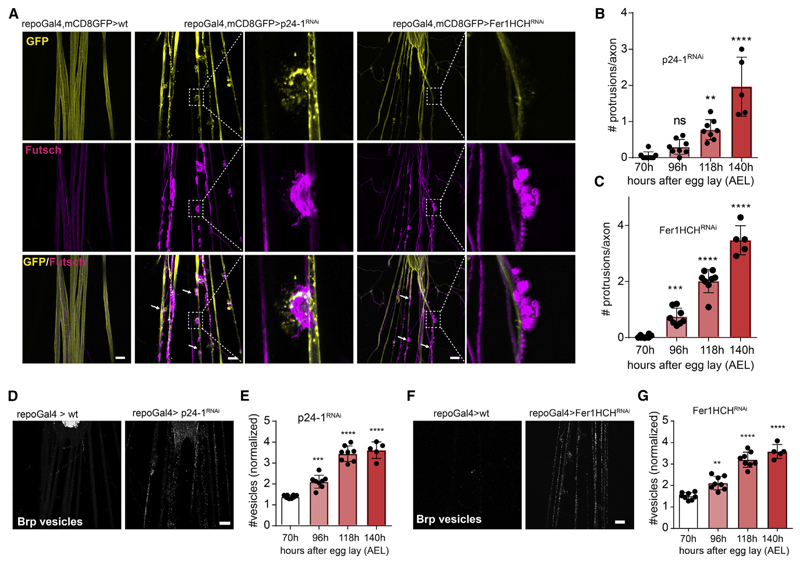
Figure 1. A Drosophila Screen Identifies a Function for p24-1 and Ferritin 1 Heavy Chain in Maintaining Axonal Transport and Integrity.
(A) Images showing third instar Drosophila melanogaster larva peripheral nerves with repoGal4-driven CD8-GFP to visualize glia (yellow) and immunostained against the microtubule-associated protein futsch (magenta) to label axons. p24-1 and ferritin 1 heavy chain homolog (Fer1HCH) knockdown in glia using re-poGal4 results in nerve damage with the formation of focal protrusions. White arrows indicate focal axonal degenerations. Insets show higher magnifications of protrusions. Scale bar, 20 μm.
(B and C) Quantification of number of axonal protrusions per nerve afterglial-specific RNAi knockdown of p24-1 (B) and Fer1HCH (C) at second instar(70 h after egg lay [AEL]), mid third instar (96 h AEL), late third instar (118 h AEL), and prolonged third instar larval stages (140 h AEL).
(D) Immunostaining with antibodies against Bruchpilot (Brp) shows the accumulation ofBrp-positivevesicles after glial-specific knockdown of p24-1 in third instar larvae. Scale bar, 40 μm.
(E) Quantification of Brp-positive vesicles per nerve after glial-specific RNAi knockdown of p24-1 in second instar (70 h AEL), mid third instar (96 h AEL), late third instar (118 h AEL), and prolonged third instar larval stages (140 h AEL).
(F) Immunostaining with antibodies against Brp shows the accumulation of Brp-positive vesicles after glial-specific knockdown of Fer1HCH in third instar larvae as indicated. Scale bar, 20 μm.
(G) Quantification of Brp-positive vesicles per nerve afterglial-specific RNAi knockdown of Fer1HCH in second instar(70 h AEL), mid third instar (96 h AEL), late third instar (118 h AEL), and prolonged third instar larval stages (140 h AEL).
In (E) and (G), the values are normalized to mean of 70 h. All data are means ± SD; ns, non-significant; **p < 0.01, ***p < 0.001, ****p < 0.0001 by one-way ANOVA with Dunnett’s multiple comparison test.