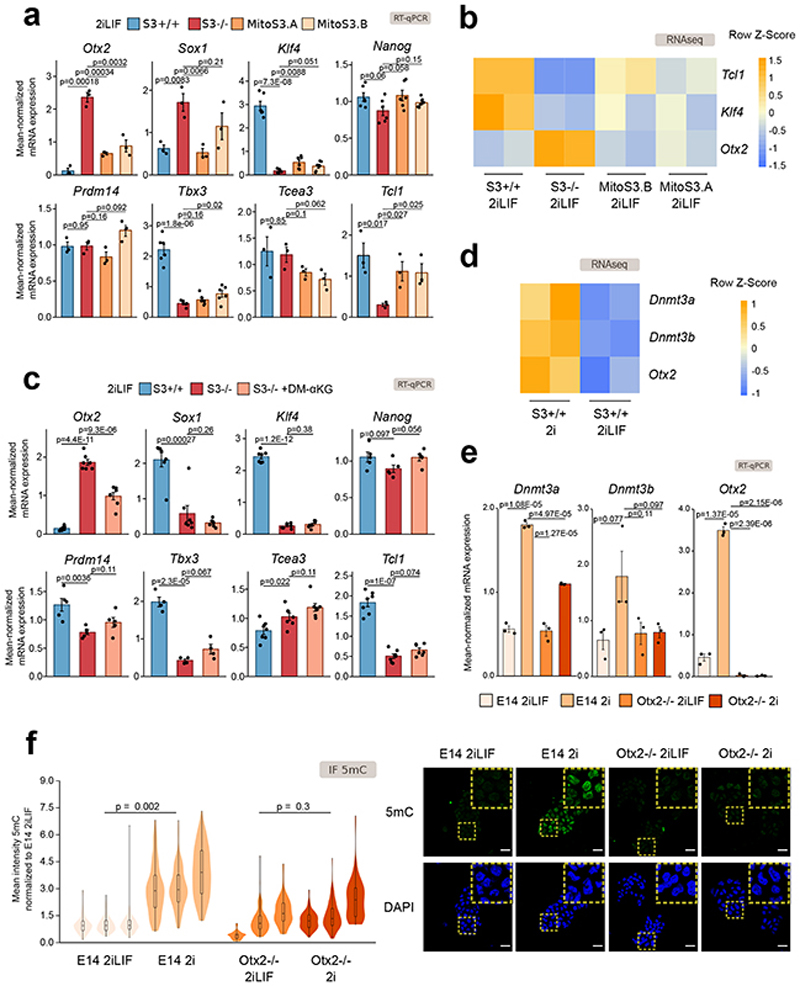
Fig. 6. Otx2 links α KG to Dnmt3a/b expression.
a-b, Expression analysis of S3+/+, S3-/-and MitoS3.A/B clones cultured in 2iLIF for potential Dnmt3a/b regulators.
a, RT-qPCR. Bars: mean ±s.e.m. of n = 3 for Otx2, Sox1, Prdm14, Tcea3, Tcl1; n = 6 for Klf4, Nanog, Tbx3 experiments, shown as dots.
b, Heatmap of RNA-seq data; n = 2 biological replicates. Z-scores of scaled expression values.
c, RT–qPCR of S3+/+, S3-/- and S3-/- cells cultured in 2iLIF and treated with 2 mM DM-αKG for 3 passages. Bars: mean ±s.e.m. of n = 7 for Otx2, Sox1, Klf4, Tcea3, Tcl1; n = 5 for Nanog, Prdm14; n = 4 for Tbx3 experiments, shown as dots.
d, Heatmap reporting expression of Dnmt3a, Dnmt3b and Otx2 in S3+/+ cultured in 2i with or without LIF; n = 2 biological replicates. Expression levels were scaled and represented as z-score.
e, RT–qPCR of E14 and Otx2-/- cells stably cultured in 2iLIF or 2i. Bars: mean ±s.e.m. of n = 3 experiments, shown as dots.
f, Anti-5mC immunofluorescence on E14 and Otx2-/- cells stably cultured in 2iLIF or 2i. Representative images and violin plots of fluorescence intensity of an average of 111 nuclei per sample. Boxplots show 1st, 2nd and 3rd quartile; Whiskers indicate minimum and maximum values. n = 3 experiments.
All P values calculated using two-tailed unpaired t-test. Scale bar: 20 μm.