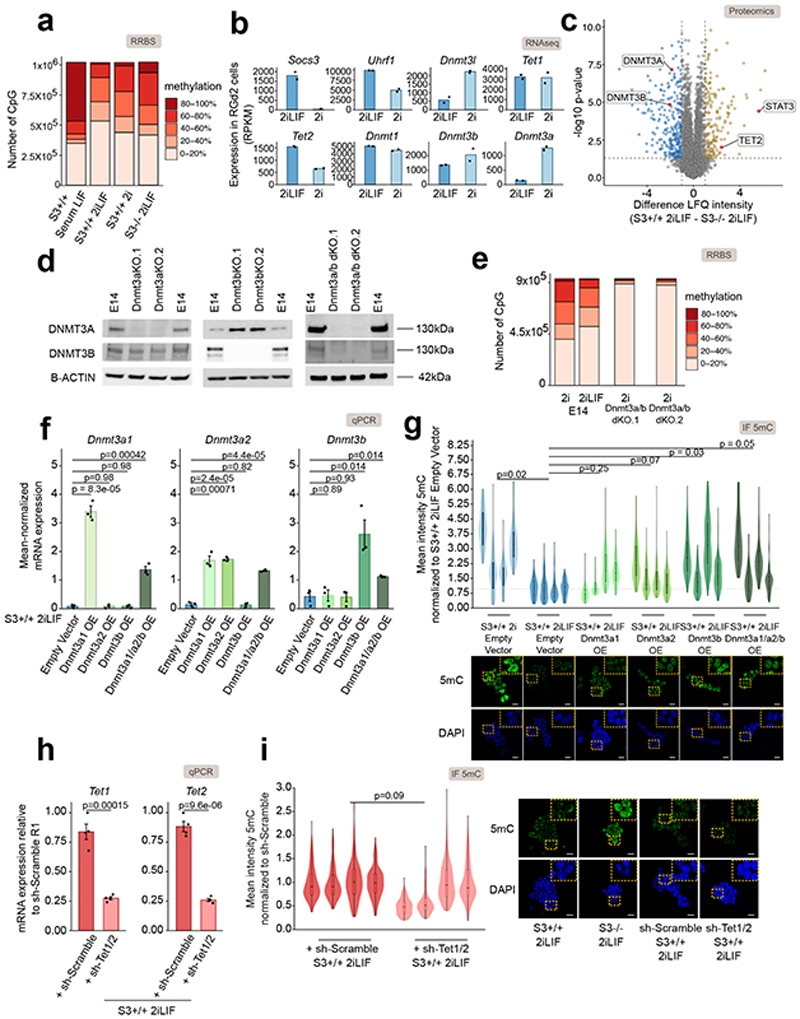
Extended Data Fig. 1. Dnmt3a/b controls 5mC levels downstream of LIF/Stat3.
a, Distribution of DNA methylation levels at CpG islands in S3+/+ cells cultured in Serum LIF, 2i or 2iLIF and S3-/- cells in 2iLIF.
b, Gene expression analysis in RGd2 cells in 2i with or without LIF. Socs3 was used as a control of LIF/Stat3 activation. Bars: mean of n=2 biological replicates.
c, Proteomics data from S3+/+ and S3-/- cells in 2iLIF. Yellow and blue dots indicate respectively proteins that are more or less abundant (difference > 1 or < -1, p-value < 0.05) in S3+/+ relative to S3-/-. n=5 biological replicates.
d, Western blot of E14 mES cells and of Dnmt3a KO, Dnmt3b KO, and Dnmt3a/b double KO cells (two clones for each mutant genotype) in Serum LIF. Two E14 samples are loaded on the right and left for each KO cell line. B-ACTIN used as a loading control. Representative images of n=2 independent experiments.
e, Distribution of DNA methylation levels at CpG islands in E14 mES cells in 2i or 2iLIF and two clones of Dnmt3a/b double KO mES cells in 2i.
f, Gene expression analysis of S3+/+ cells cultured in 2iLIF transiently expressing an Empty Vector, Dnmt3a (two isoforms, Dnmt3a1 and Dnmt3a2 – as previously identified in72), Dnmt3b, or the three genes simultaneously (Dnmt3a1/a2/b OE). Bars: mean ± s.e.m. of n=3 independent experiments, shown as dots.
g, Anti-5mC immunofluorescence on S3+/+ in 2i and 2iLIF, and Dnmt3a1 OE, Dnmt3a2 OE, Dnmt3b OE, and Dnmt3a1/a2/b triple OE cells in 2iLIF. Violin plots of an average of 89 nuclei per sample. n=4 experiments.
h, Gene expression analysis of S3+/+ cells in 2iLIF stably expressing shRNA to knock-down Tet1 and Tet2 simultaneously or a scrambled control shRNA. Bars: mean ±s.e.m. of n=4 experiments.
i, Anti-5mC immunofluorescence on S3+/+ in 2iLIF transiently expressing a scrambled control shRNA and shRNA against Tet1/Tet2. Violin plots of an average of 78 nuclei per sample. n=4 experiments.
All violin and boxplots indicate the 1st, 2nd and 3rd quartiles, with whiskers indicating minimum and maximum value. All p-values calculated by two-tailed unpaired T-test. Scale bars: 20μm.