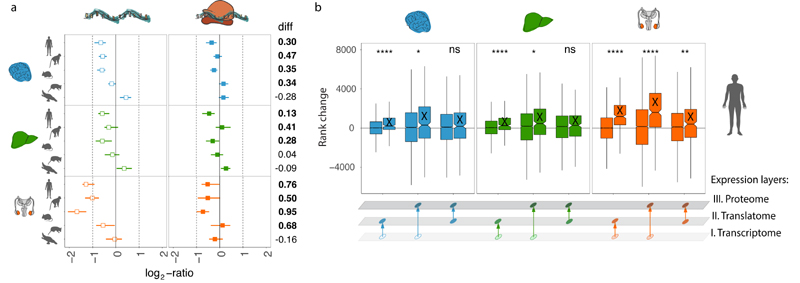
Fig. 2. Evolution of gene expression across expression layers.
a-c, Gene expression phylogenies of 5,060 robustly expressed (FPKM > 1 across all libraries) 1:1 orthologues at the transcriptome (light and thick branches) and translatome (dark and thin) layers for brain (a), liver (b), and testis (c). Branch lengths represent the fractions of expression variation, which correspond to evolutionary changes in expression levels (Extended Data Fig. 5a). Due to the lack of a biological replicate, the branch leading to human was omitted in the liver phylogeny for the transcriptome layer. Proportions of bootstrapped trees supporting branching patterns are indicated next to the respective nodes. d-f, Differences in the evolution between transcriptome and translatome layers for individual genes in brain (d), liver (e), and testis (f). Density distribution, median Δ, interquartile range (IQR) of Δ, and number of cases with Δ significantly higher (potentially driven by directional selection) or lower (stabilizing selection) than zero are shown to the right of each panel. All genes in graphs d-f can be interactively explored in our Ex2plorer database (https://ex2plorer.kaessmannlab.org/). g, Similarity of gene expression (rank) changes between human and mouse brains at the proteome layer compared to the changes at the underlying translatome and transcriptome expression layers, respectively, as assessed by Spearman’s correlation coefficients (ρ). Proteomics data were retrieved from previous studies21,22. Organ and species icons were previously used in ref. 25.