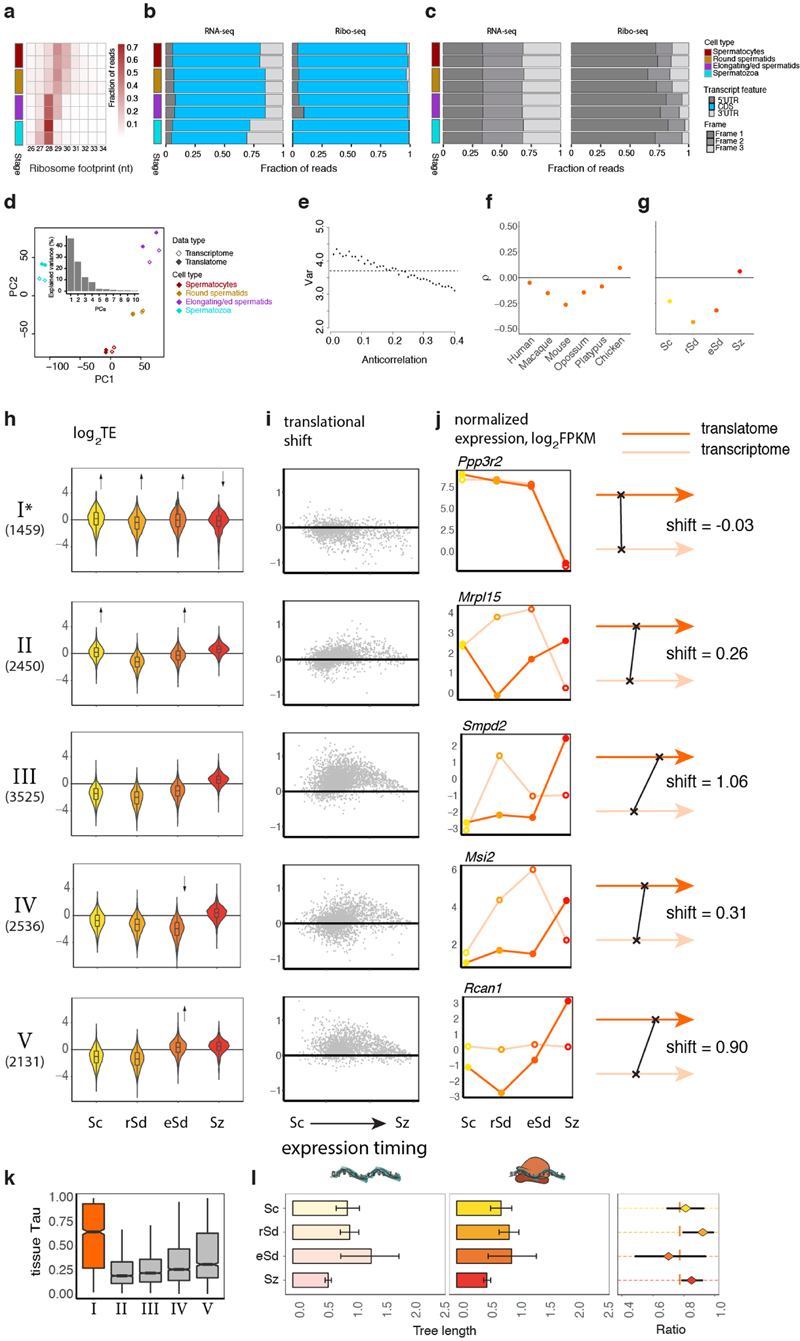
Extended Data Fig. 3. Quality assessment and analysis of mouse spermatogenesis data.
a, Ribosome footprint length distributions across Ribo-seq libraries (nt, nucleotides). b, Fractions of Ribo-seq and RNA-seq reads mapped to 5’ untranslated regions (5’UTRs), coding sequences (CDS), and 3’ untranslated regions (3’UTRs), respectively. c, Distribution of Ribo-seq and RNA-seq reads across the three reading frames in the coding sequence (CDS) of dominant splicing isoforms (Frame 1: canonical reading frame). d, PCA based on 11,057 genes robustly expressed (median FPKM > 1) across murine spermatogenesis libraries. The scree plot (inset) indicates the percentage of variance explained by each of the first 10 PCs. e, Variance at the translatome layer calculated for simulated scenarios with different amounts of translational contribution (see Methods for details). Dashed line corresponds to IQR calculated at the transcriptome layer; f,g, Spearman’s ρ between transcription abundance and TE was calculated for 5,060 robustly expressed (median FPKM > 1 across organ libraries) 1:1 amniote orthologues in bulk testis across the amniotes (f) and across spermatogenesis stages in mouse (g). h,i, TE (h) and translational shift (i) for clusters of genes (gene numbers in parentheses) with distinct TE patterns (Mfuzz clustering). Arrows indicate TE increases/decrease compared to the respective global pattern (Fig. 1e). *indicates a cluster of genes, which escape expression repression and delay at the translatome layer. j, Expression of individual genes, representing each of the five TE clusters, at the transcriptome and translatome layers (left column); shift in expression timing between expression layers for the corresponding genes (right column) with crosses representing the centers of mass of gene expression across spermatogenesis. k, Tissue-specificity (tissue Tau) across TE clusters. Cluster I, highlighted in color, is dominated by testis-specific genes. Box plots represent the median ± 25th and 75th percentiles, whiskers are at 1.5 times the interquartile range. l, Gene expression divergence at the two expression layers for genes with stage-specific expression across spermatogenesis among 8,109 1:1 orthologues robustly expressed (FPKM > 1) in macaque, mouse, and opossum. Sc, spermatocytes, rSd, round spermatids, eSd, elongating/elongated spermatids, Sz, spermatozoa.