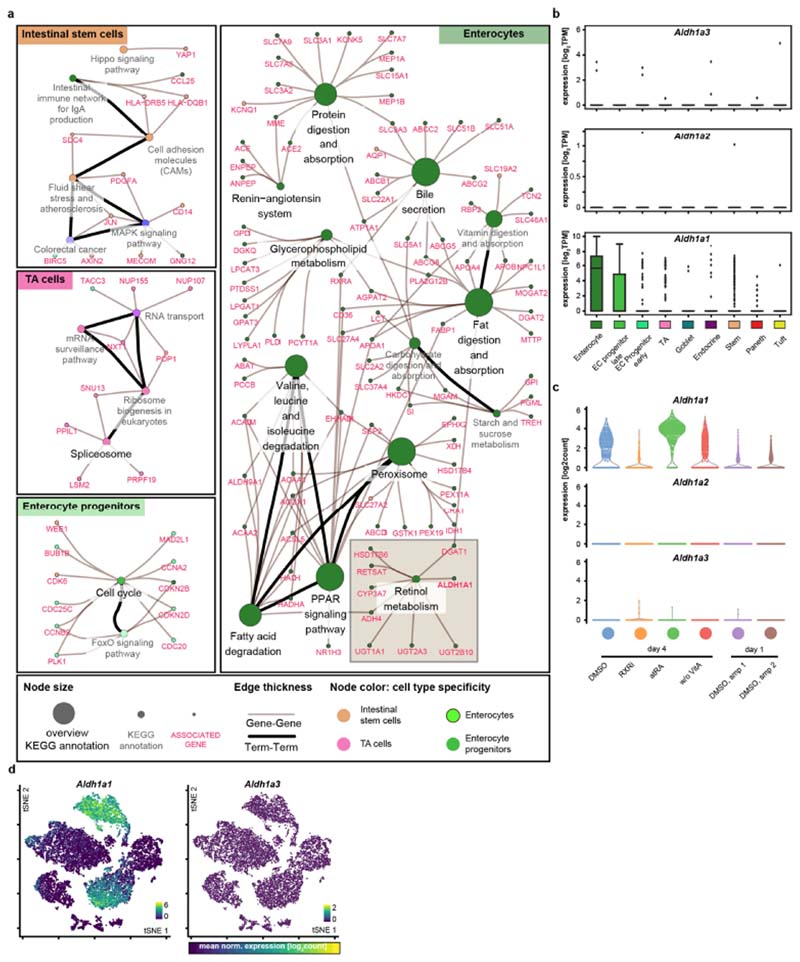
Extended Data Fig. 6. Retinol metabolism in enterocytes.
a, Network representation of annotation enrichment analysis for a published RNA-seq dataset describing marker genes of intestinal epithelium cell types29. Small nodes represent genes; large nodes represent functional annotations; edge thickness show gene-annotation assignments (grey) or term–term relations (black). Colours indicates cell-type specificities of annotations (see key). b, Box plots showing expression (in log2-normalized transcripts per kilobase million (TPM) values) of Aldhlal, Aldh1a2 and Aldh1a3 across cell types in the published single-cell RNA-seq (scRNA-seq) dataset29. Boxes show quartile ranges; whiskers represent value intervals with excluded outliers; middle lines show median values. Each data point represents a single cell. EC, enterocyte; TA, transient-amplifying. n = 1,522 single cells. c, Expression (in log2 normalized transcript counts) of Aldhlal, Aldh1a2 and Aldh1a3 across indicated treatment conditions in scRNA-seq (Fig. 3 and Extended Data Fig. 9). Sampling of n = 3,000 single cells. d, tSNE maps of the scRNA-seq experiment. Each data point represents a single cell, colour-coded by expression (log2 normalized transcript counts) of indicated genes in samples of the scRNA-seq experiment (Fig. 3). Ranges of the colour mapping are indicated in each plot. Sampling of n = 5,000 single cells. b–d, Each data point represents a single cell. atRA, 10 μM all-trans retinoic acid; DMSO, DMSO control, RXRi, 5 μM RXR antagonist; w/o VitA, vitamin-A-depleted medium.