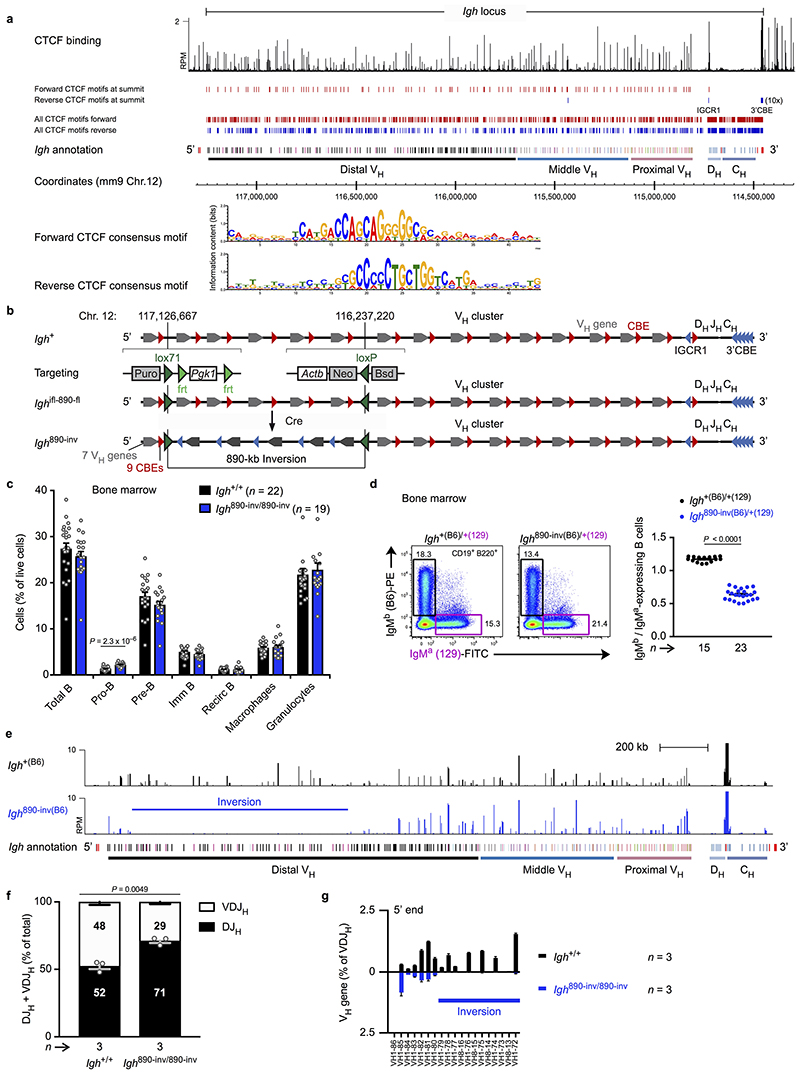
Extended Data Fig. 1. Generation and characterization of the Igh 890-inv allele.
a, Orientation of the CTCF-binding sites in the Igh locus. The CTCF-binding pattern was determined by ChIP-seq of Rag2 –/– pro-B cells. The locations of forward (red) and reverse (blue) CTCF motifs detected at the summit of the CTCF peaks are shown together with all predicted CTCF motifs, identified based on the forward and reverse consensus CTCF-binding motifs shown. The annotation of the C57BL/6 Igh locus indicates the distinct VH gene families (different colours) in the distal, middle and proximal VH gene regions (Johnston et al. 2006. J. Immunol. 176, 4221-4234) and the 3’ proximal Igh domain containing the DH, JH and CH elements as well as the Eμ and 3’RR enhancers (red). b, Schematic diagram of the Igh ifl-890-fl and Igh 890-inv alleles. The indicated selection cassettes were used for introducing the upstream inverted lox71 site (ifl) and downstream loxP site (fl) by sequential ES cell targeting. c, Flow cytometric analysis of bone marrow cells from Igh 890-inv/890-inv and Igh +/+ mice. The relative frequencies of the indicated cell types (defined in Methods) are shown as mean values with SEM. d, Flow cytometric determination of the ratio of immature IgMb (B6) to IgMa (129) B cells from Igh 890-inv(B6)/+(129) and Igh +(B6)/+(129) mice, which were generated by crossing Igh 890-inv/+ mice on the C57BL/6 (B6) background with Igh +/+ mice of the 129/Sv (129) strain. The rearranged Igh alleles of the C57BL/6 and 129/Sv strains give rise to expression of the IgMb and IgMa isotypes, respectively. e, VH gene expression from the Igh 890-inv(B6) or Igh +(B6) allele in immature IgMb (B6) B cells sorted from Igh 890-inv(B6)/+(129) or Igh +(B6)/+(129) mice, respectively. The RNA-seq profiles are shown with the Igh annotation (see a) and inverted region (bar). One of two experiments is shown. RPM, reads per million mapped sequence reads. f,g VDJ-seq analysis of pro-B cells from the bone marrow of Igh 890-inv/890-inv and Igh +/+ mice. f, The percentages of uniquely identified DJH and VDJH sequences are indicated. g, The relative usage of the distal VH genes at the Igh 5’ end is shown as mean percentage of all VDJH recombination events with SEM. A horizontal bar indicates the 5’ end of the inverted region. Statistical data are shown as mean value with SEM and were analysed either by multiple t-tests (unpaired and two-tailed with Holm-Sidak correction; c,f) or by the Student’s t-test (unpaired and two-tailed; d). n, number of mice (c,d) or experiments (f,g). Each dot corresponds to one mouse.