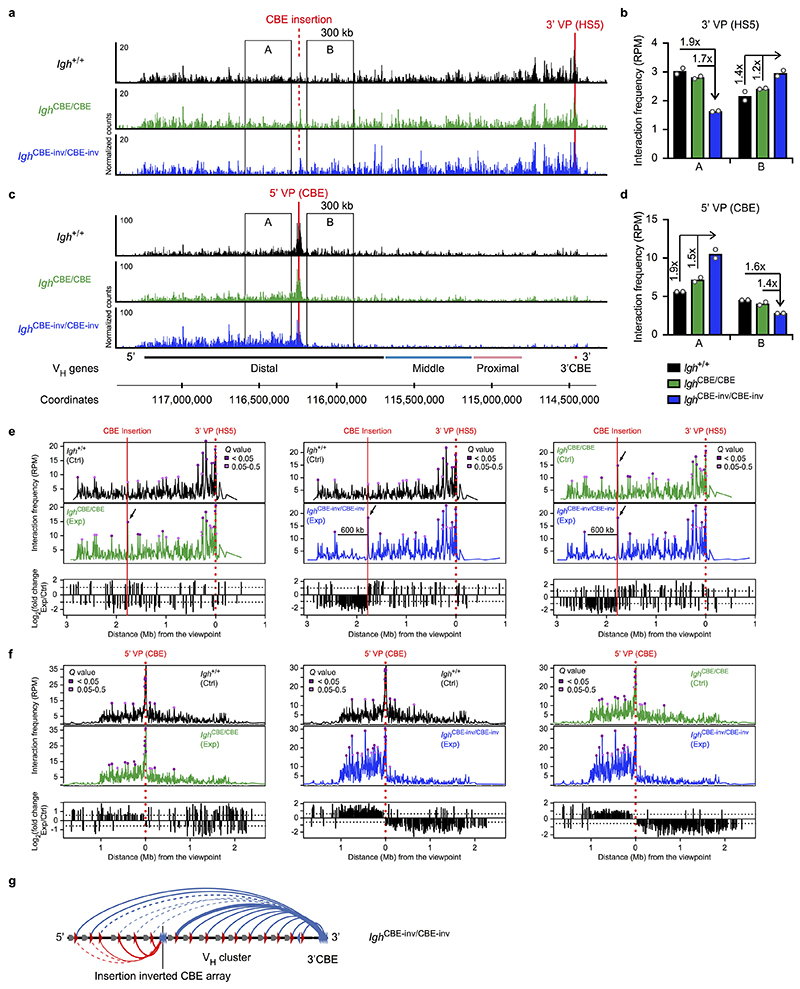
Extended Data Fig. 4. New loop formation upon insertion of an inverted CBE array in the Igh locus.
a-d, 3C-seq analysis of interactions across the Igh locus from the 3’ (HS5; a,b) or 5’ (CBE; c,d) viewpoint in short-term cultured Igh CBE-inv/CBE-inv , Igh CBE/CBE and Igh +/+ pro-B cells. a,c, Mapping of the 3C-seq reads as normalized counts across the Igh locus. One of two experiments per genotype is shown. b,d, Quantification of the interactions from the 3’ (HS5, b) or 5’(CBE, d) viewpoint. The 3C-seq reads in regions A and B were quantified as mean RPM value per region, based on two experiments per genotype. e,f, Interactions from the 3’ (HS5; e) or 5’ (CBE; f) viewpoint in Igh +/+, Igh CBE/CBE and Igh CBE-inv/CBE-inv pro-B cells. The 3C-seq reads along the Igh locus (upper two panels) are shown as RPM values with the respective Q values, calculated by the r3Cseq program based on two experiments per genotype. Arrows highlight a strong interaction from the 3’ viewpoint (HS5) to a region immediately downstream of the inserted CBE array (e), and a bar denotes the 600-kb region exhibiting reduced interactions in Igh CBE-inv/CBE-inv pro-B cells (e). The lower panel shows the fold-change of the RPM values for interaction differences of > 2-fold (dashed line; e) or > 1.5-fold (f) between the experimental (Exp) and control (Ctrl) pro-B cells of the indicated genotypes. g, Schematic diagram summarising the loop formation at the Igh CBE-inv allele in pro-B cells. The CBEs with their orientation are indicated by red and blue arrowheads, VH genes by grey arrows and loops by arches. The new loop domain is indicated in red. A thicker blue arche indicates the enhanced interaction from the 3’CBE to a region immediately downstream of the inserted CBE array.