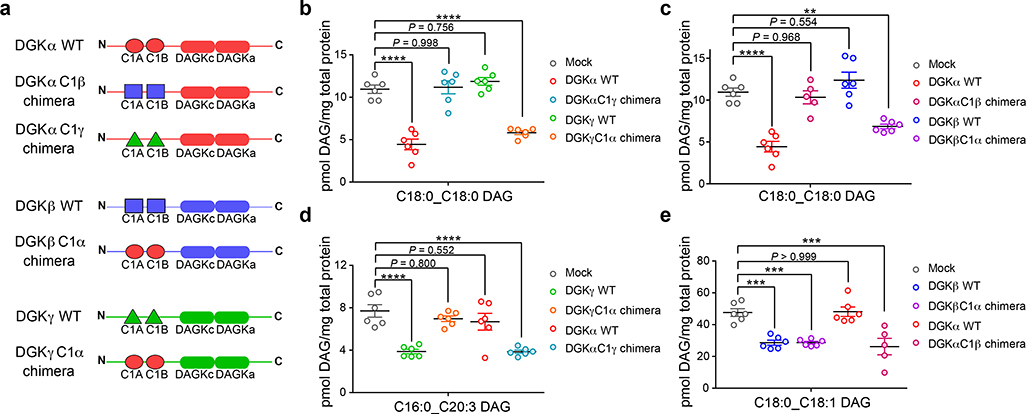
Figure 5. Programming DAG substrate specificity of DGKs in live cells by C1 domain engineering.
a) Schematic of recombinant rat DGK wild-type (WT) and chimeric proteins used for metabolomics analyses. See Supplementary Note for plasmid construct sequences and Supplementary Fig. 18 and 19 for validation of chimeric protein expression and activity. Targeted lipidomics demonstrated that DAG fatty acyl specificity of type 1 DGKs can be engineered via C1 domain swapping: b) DGKα C18:0_C18:0 DAG specificity transferred to DGKγ; c) DGKα C18:0_C18:0 DAG specificity transferred to DGKβ; d) DGKγ C16:0_C20:3 DAG specificity transferred to DGKα; e) DGKβ C18:0_C18:1 DAG specificity transferred to DGKα. Significance of lipid species alterations was determined using a Tukey’s multiple comparison correction following a one-way ANOVA test (**P < 0.01, ***P < 0.001, ****P < 0.0001). All data show represents mean ± s.e.m.; (n=5–6 biological samples). All data shown are representative of two experiments (n=2 biologically independent experiments).