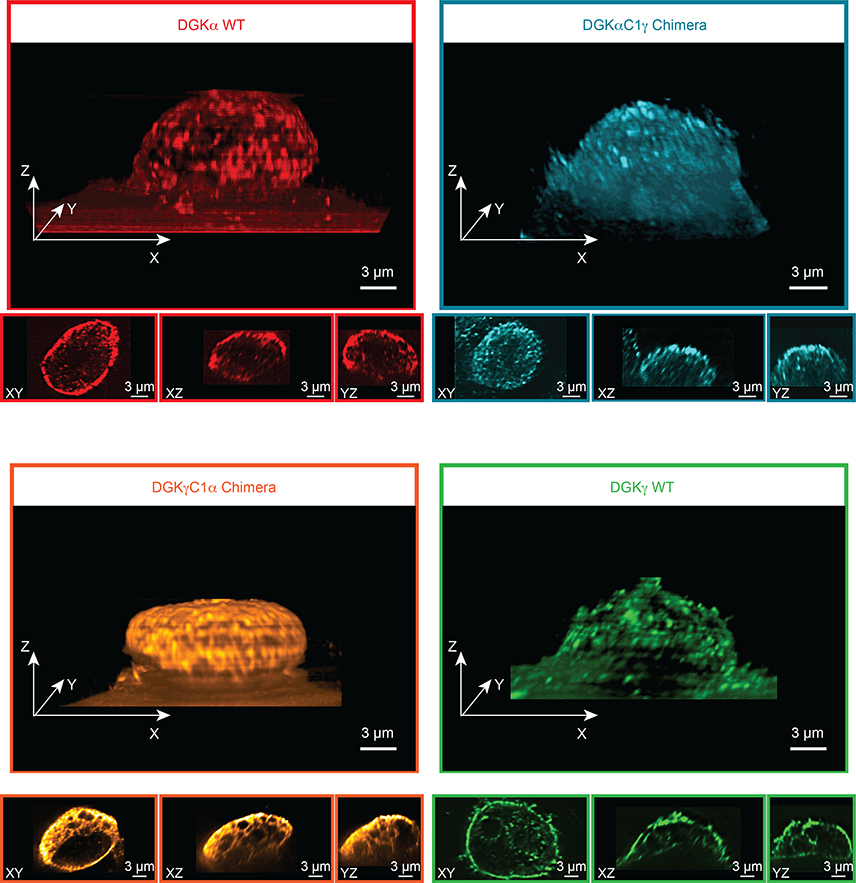
Figure 6. Lattice-light sheet microscopy shows DGK chimeras are not localized based on C1 identity.
Recombinant rat DGK isoforms were imaged by immunofluorescence using an anti-FLAG primary and DyLight 550 secondary antibody staining. Images were acquired at 561 nm excitation using an illumination intensity of the sample at 1 W/cm2. The raw 3D image data was de-skewed and deconvolved as described previously45, 46 and 3D images were rendered using the 3D Viewer plugin in Fiji (see Methods for details). The reconstructed voxel size of the 3D image is 100 × 100 × 100 nm. For each 3D image, three orthogonal cross sections through the cell center are also displayed. Microscopy results reveal that subcellular localization of the C1 chimeras are different from each other and from wild-type C1 donor DGKs. Data shown are representative of two experiments (n=2 biologically independent experiments).