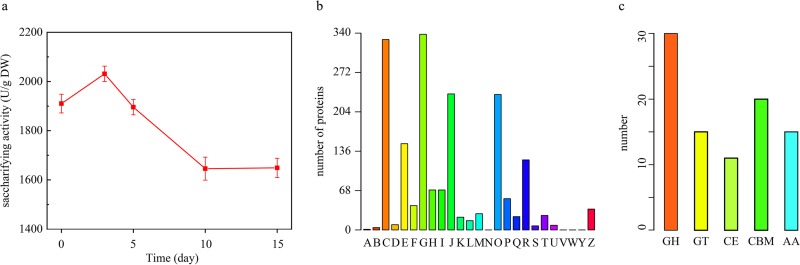
FIG 1.
Enzyme profile in baijiu fermentation. (a) Dynamic of saccharifying activity in baijiu fermentation. (b) Identified protein functions in COG database annotation. A, RNA processing and modification; B, chromatin structure and dynamics; C, energy production and conversion; D, cell cycle control, cell division, and chromosome partitioning; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; G, carbohydrate transport and metabolism; H, coenzyme transport and metabolism; I, lipid transport and metabolism; J, translation, ribosomal structure, and biogenesis; K, transcription; L, replication, recombination, and repair; M, cell wall/membrane/envelope biogenesis; N, cell motility; O, posttranslational modification, protein turnover, and chaperon functions; P, inorganic ion transport and metabolism; Q, secondary metabolites biosynthesis, transport, and catabolism; R, general function prediction only; S, function unknown; T, signal transduction mechanisms; U, intracellular trafficking, secretion, and vesicular transport; V, defense mechanisms; W, extracellular structures; Y, nuclear structure; Z, cytoskeleton. (c) Category of identified carbohydrate hydrolases in the baijiu fermentation. GH, glycoside hydrolase families; GT, glycosyltransferase families; CE, carbohydrate esterase families; AA, auxiliary activity families; CBM, carbohydrate-binding module families. Bar, numbers of proteins.