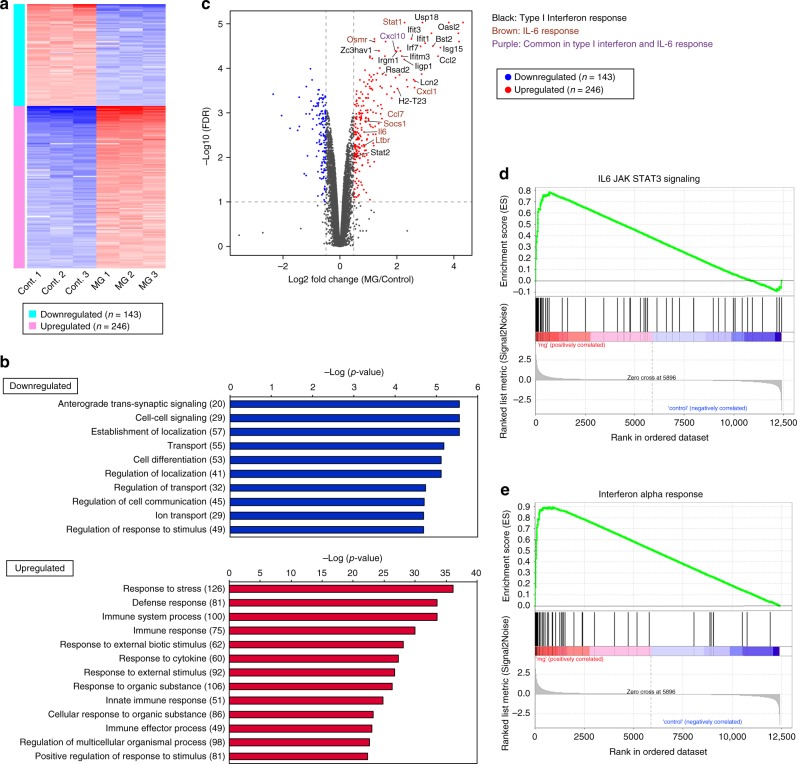
Fig. 8. Signaling pathways triggered by IL-6 and IFN-I were activated in CP-like neurons after coculture with microglia.
a Transcriptomic profiles of the neuronsCont and neuronsMG groups based on differentially expressed gene (DEG) analysis. Among the genes that were reliably detected in both groups (11,603 genes), we identified 143 downregulated genes and 246 upregulated genes in neuronsMG group based on the statistical threshold (log2 fold change [FC] > 1.4, FDR < 0.1). b Gene ontology (GO) analysis for significantly downregulated and upregulated genes (see Supplementary Table 3). c A volcano plot depicting DEGs between the neuronsCont and neuronsMG groups. The negative log of FDR is plotted on the Y-axis, and the log2 FC of neuronsMG/neuronsCont is plotted on the X-axis. The blue and red points on this graph represent mRNAs that were significantly downregulated (n = 143) and upregulated (n = 246), respectively, in neuronsMG (FC > 1.4; FDR < 0.1). The color of the gene names indicates that the genes are typical in the type I interferon response (black), typical in the IL-6 response (brown), and common in type I interferon and IL-6 responses (purple). Gene set enrichment analysis showing a barcode plot for genes registered in IL-6 JAK STAT3 signaling (d) or the interferon alpha response (e). Genes were ranked based on their differential expression between neuronsMG and neuronsCont. A plot of running enrichment score (RES) is shown in green (top). The vertical black bars (middle) indicate a gene within each gene set. The correlation of the expression pattern of genes belonging to each subcluster is shown in the gray histogram (bottom). The RNA-Seq data are summarized in Supplementary Data 1.