Table 1.
Effect of pyrazineguanidine and benzoylguanidine compounds on viability of MCF-7 and MDA-MB-231 spheroids.
| Structurea | Lipophilicityb | Reported Ki values for NHE1 (µM)c | Reported IC50 values for NHE1 (µM)d | Experimentally determined EC50 values for compound-induced loss of viability (µM)e | ||||
|---|---|---|---|---|---|---|---|---|
| MCF-7 | 95% CI | MDA-MB-231 | 95% CI | |||||
| Amiloride | 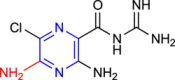 |
0.68 (LogD) 0.93 (LogP) | 1.0–7.0 | 3.7–60 | 30.5 | 18.23–42.28 | 55.8 | 32.51–81.68 |
| DMA |  |
1.64 (LogD) | 0.023–0.5 | 0.19 | 16.6 | 8.47–32.41 | 59 | 30.43–114.20 |
| EIPA | 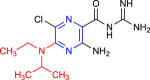 |
3.2 (LogD) | 0.01–0.1 | 0.016–0.8 | 4.8 | 1.29–17.78 | 19.6* | - |
| HMA |  |
2.94 (LogD) | 0.013 | — | 2.5 | 0.86–7.33 | 13.5 | 4.73–38.34 |
| Cariporide | 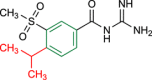 |
−1.01 (LogP) | 0.008–0.04 | 0.03–3.4 | ND** | ND | ND** | ND |
| Eniporide | 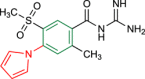 |
−1.03 (LogP) | — | 0.0045–0.38 | ND*** | ND | ND*** | ND |
The table displays reported IC50 and Ki values for NHE1 for the tested inhibitors, alongside experimentally obtained EC50 values for compound-induced loss of viability. aChemical structure of inhibitors. Blue: pyrazine ring characteristic for pyrazineguanidine compounds. Green: phenyl ring characteristic for benzoylguanidine compounds. Red highlights the different substitutions on the 5-position. bThe reported lipophilicity of each compound indicated by LogD and/or LogP values. cReported NHE1 Ki values presented in µM. dReported NHE1 IC50 values, determined in the presence of 100–130 mM Na+, presented in µM. eExperimentally obtained EC50 values based on 3 n for each inhibitor. EC50 values were calculated in GraphPad Prism by nonlinear regression using the equation Y = Bottom + (Top-Bottom)/(1 + 10^((X-logIC50))), where X is the logarithm of the inhibitor concentration, top and bottom are the highest and lowest cell viability responses measured, respectively, and Y is the relative cell viability. The corresponding 95% confidence intervals for each EC50 value is presented alongside. ND: not determined. *EC50 calculated from trendline equation generated in Excel. **No effects of concentrations up to 10 µM. ***No effect of concentrations up to 20 µM. Table refs. 13,15,17,42,48–51,56–61.
