Fig. 3.
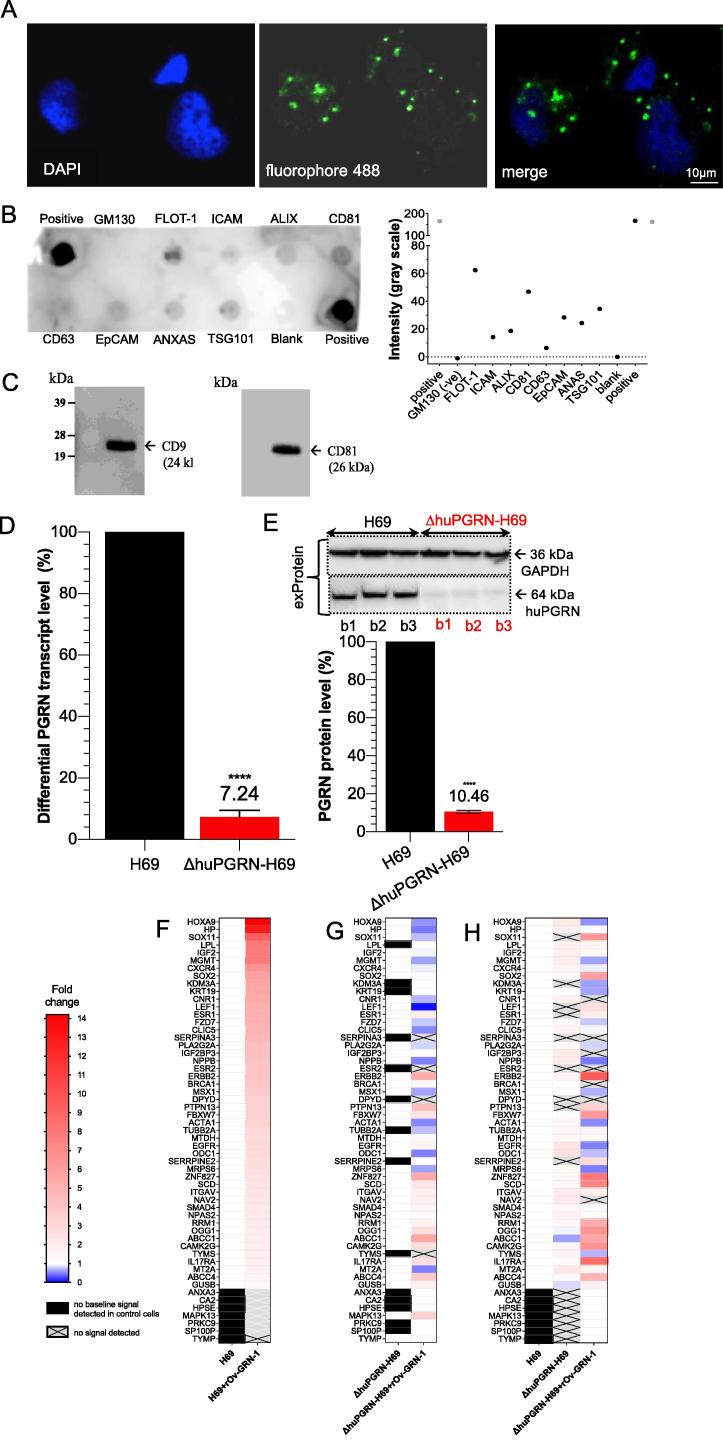
EV-derived H69 cell characterization and PGRN expression. Panel A, the visualization of EVs from H69 cells by in situ hybridization using fluorophore 488-labeled anti-CD81 antibody (green dot, panel A) around DAPI-stained cell nuclei (blue color). The H69-derived EV-like particle sizes ranged from 55 to 80 nm. The protein composition of EVs were determined using the Exosome Antibody Array (SBI System Biosciences). Panel B, dark spots indicate presence of the target protein (upper panel). Absence of a spot for GM130 confirmed the absence of cellular contaminants. The intensity in gray scale was also plotted (lower panel). Panel C, WB analysis for the exosome-specific markers; strongly positive signal for CD9 and CD81 were found. Panels D and E, both the evRNA and evProtein from ΔhuPGRN-H69 showed reduction of ∼90% differential transcript levels [red bar, panel D] after normalization with GAPDH in comparison to H69 [black bar] (red bar, panel D). The levels of progranulin were reduced by ∼90%; unpaired t-test, P < 0.0001 (****), n = 3. Panels F–H, show an induction of CCA-related mRNAs carrying cholangiocyte derived-EVs after exposure to liver fluke granulin. The heat map was plotted based on the differential transcript fold change as determined using the CCA-related gene Array using H69 as the reference from H69 with liver fluke granulin treatment (panel F) and ΔhuPGRN-H69, ΔhuPGRN-with liver fluke granulin treatment (panel H). Similar quantities of evRNAs from each group were probed individually with 88 CCA targets in triplicate; there were slight differences (P < 0.05 by 2way ANOVA gene profiles (55 from 88 transcripts were detected) in ΔhuPGRN-H69 vs H69 (panel A) by absent of DPYD, ESR, KDM3A, LEF1, PRKC9, PTPN13, SERPINA3, SERRPINE2 and SOX11 (panel H). However, most of these genes were recovered after exogenous liver fluke granulin treatment of ΔhuPGRN-H69 (panels G and H). The CCA mRNA profiles after liver fluke granulin treatment of EVs derived from ΔhuPGRN-H69 and H69 were markedly dissimilar following exposure to liver fluke granulin with P < 0.0001, 2way ANOVA (panels F and G). Panel G, shows the CCA-related evRNAs after exposure to liver fluke granulin in ΔhuPGRN-H69 compared with its mRNA profile (P < 0.0001 with two-tailed, paired t-test). The statistical significance was calculated from differential fold changes in transcription. The liver fluke granulin (rOv-GRN-1) treatment group of cells showed 1–15 folds (gradient red bar) level of induction of transcription. The ‘1’ indicates baseline (white bar) level expression, and ‘0’ indicated the absence of expression (gradient blue bar). The transcripts from non- or treated- rOv-GRN-1treatment were not detected (no baseline signal) indicate in black bar and gray-cross bar, respectively. The heat map was plotted from the average transcript level ran in triplicates using Prism v8. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)