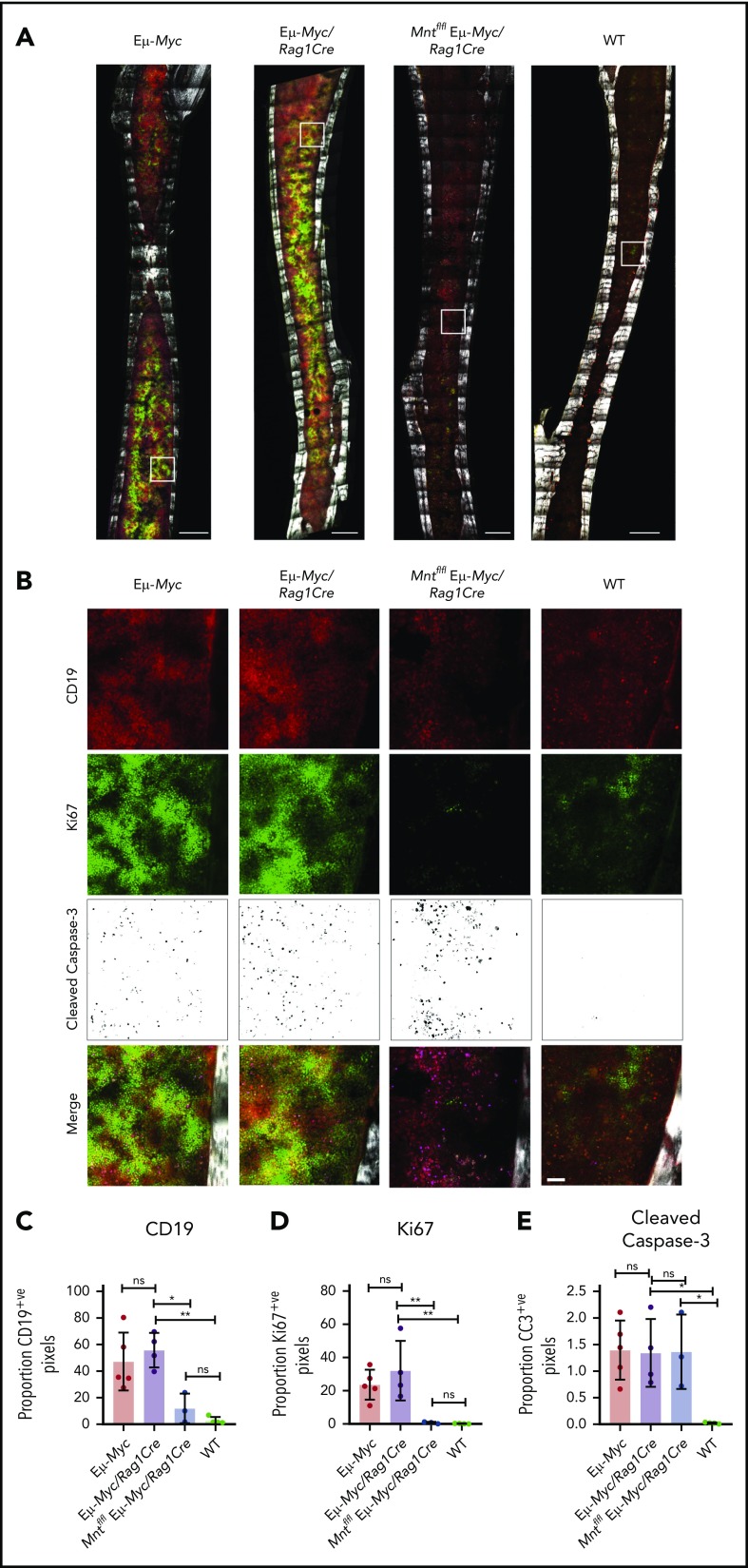
Figure 3.
In situ analysis of cell proliferation and apoptosis. (A) Tilescan sections of whole tibia. Bones were harvested from 4-week old Eμ-Myc (n = 5), Eμ-Myc/Rag1Cre (n = 4), Mntfl/fl Eμ-Myc/Rag1Cre (n = 3), and WT (n = 4) mice, stained and imaged by combined 2-photon/confocal imaging of cell populations in situ. Red = CD19; green = Ki67; magenta = cleaved caspase-3; gray = second harmonic generation (bone). To better visualize CC3-positive cells in Figure 3B, the data are presented using an inverse black and white LUT (Lookup Table). Scale bar, 500 μm. (B) Zoomed areas from original tilescans in panel A (white boxes) with individual channels for CD19, Ki67, and cleaved caspase-3. A merged image (bottom panel) from each area is shown relative to bone signal (gray). Scale bar, 50 μm. (C-E) Quantification of cells in tibia positive for CD19, Ki67, and cleaved caspase-3. Color images were separated into single binary channels and then thresholded, and the number of positive pixels for each whole bone section quantified for CD19 (C), Ki67 (D) and cleaved caspase-3 (E). Each data point represents a whole quantified bone from an individual mouse in each genotype (n = 3-5). Bar graphs show mean ± SD; significance was determined by analysis of variance *P ≤ .05; **P ≤ .01).