Figure 1.
Example Genotype Data and IBIS Processing
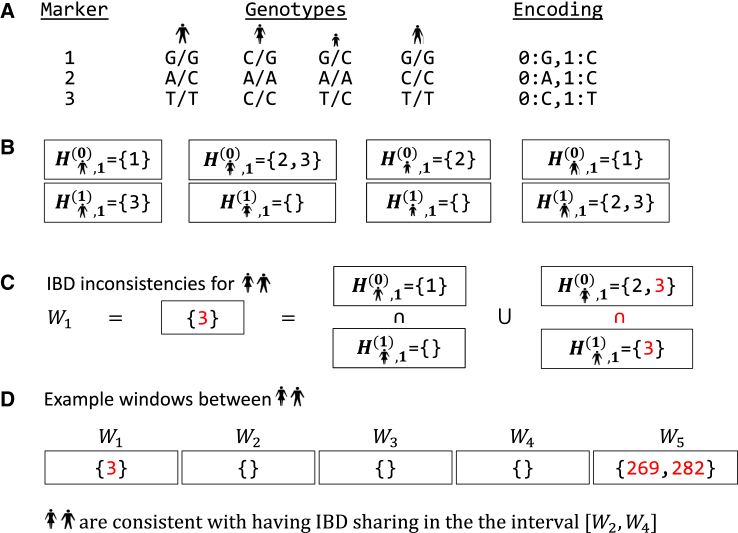
(A) Genotype data at three markers for four samples (stick figures). The input encodes the alleles at each site as a zero or one, as indicated.
(B) Sets and for each sample s contain the markers that are homozygous for alleles 0 and 1, respectively.
(C) The set operations to construct W1 for two of the samples yields one marker where these individuals are homozygous for opposite alleles.
(D) Example Wi values across five windows for a given pair. IBIS detects potential IBD segments as a series of windows where the first and last windows have no inconsistencies and where for all other windows i. Such a region with no inconsistent genotypes spans windows two through four in the figure.