Figure 3.
Transcriptomic Analysis in Human Neural Stem Cells (hNSCs) after NOVA2 Inactivation
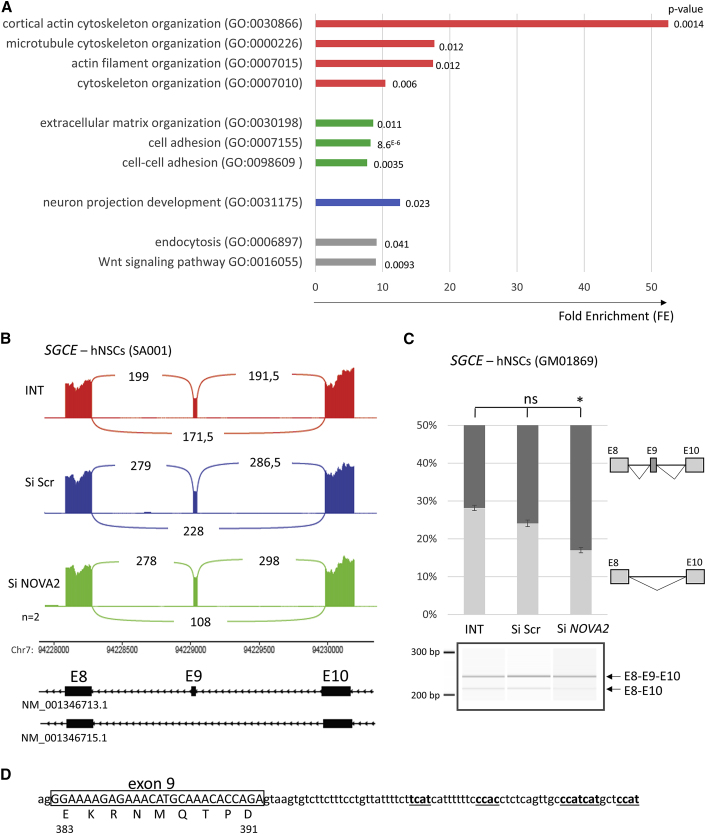
(A) Enrichment of GO terms (Biological Process and Molecular Function, analyzed using DAVID) for the genes with differential alternative splicing (AS) events identified in human neuronal precursors (hNSCs – SA001) after NOVA2 inactivation. Fold enrichement (FE) and p value are also indicated.
(B) Sashimi plot established from RNA-seq data representing the AS of SGCE exon 9 (GenBank: NM_001346713.1). The number of reads supporting the existence of each exon-exon junction is indicated as an average between data from the two independent series of hNSCs treated with INTERFERin only (in red), with scramble siRNA (in blue) or with NOVA2 siRNA (in green) during 48 h.
(C) Confirmation of the consequences of NOVA2 inactivation on SGCE splicing in another hNSC cell line (GM01869), treated with the transfecting agent only (INT) or transfected with Scramble (si Scr) or NOVA2 (siNOVA2) siRNA. The RT-PCR products obtained were analyzed by migration on a 2,100 Bioanalyzer instrument (Agilent Technology). Experiments were done in triplicates. The error bars indicate the SEM. Kruskal-Wallis’ ANOVA with Dunn’s multiple comparison test was performed ∗p < 0.05, ns: non-significant.
(D) Sequence of SGCE exon9-intron9 junction with indicated potential NOVA2 binding sequences YCAY (in bold and underlined).