Figure 2.
Disruption of Zebrafish sema6ba and sema6bb 3′ Regions Results in CNS Defects
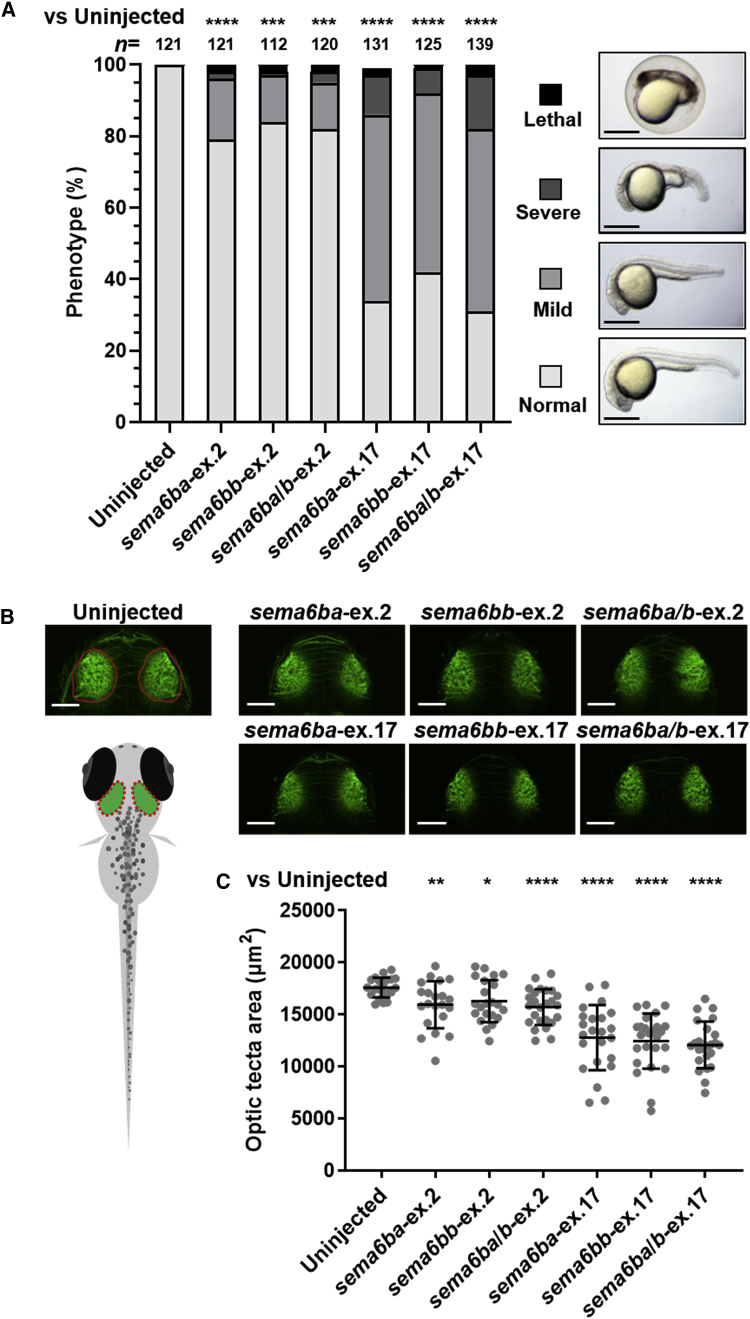
(A) Frequency of developmental defects and CNS abnormalities in embryos at 24 hpf. Zebrafish embryos were injected with the following crRNAs: sema6ba-ex.2, sema6bb-ex.2, sema6ba/b-ex.2, sema6ba-ex.17, sema6bb-ex.17, or sema6ba/b-ex.17, all of which were duplexed with tracrRNA and Cas9 protein. Uninjected embryos and F0 crispants were scored on the basis of morphological anomalies; lethal (cell death distributed over the entire body), severe (smaller head with shortened body axis), mild (developmental delay with smaller head), and normal. (n = 112–139 in each group). All experiments were performed three times. Scale bar, 500 μm. ∗∗∗p < 0.0005, ∗∗∗∗p < 0.0001 by the chi-square test.
(B) Acetylated tubulin staining of uninjected embryo and F0 crispants shows brain structures at 2.5 dpf (dorsal view). Schematic representation of neuroanatomical structures shows the area of the optic tectum (red dotted oval).
(C) F0 crispants with sema6ba-ex.17, sema6bb-ex.17, and sema6ba/b-ex.17 displayed a smaller optic tectum than uninjected embryos or sema6ba-ex.2, sema6bb-ex.2, or sema6ba/b-ex.2 F0 crispants. Scale bar, 100 μm. Data are represented as the mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.0005, ∗∗∗∗p < 0.0001 by Student’s t test.