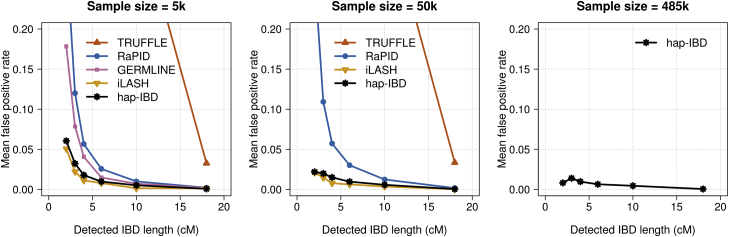
Figure 3.
False-Positive IBD Segment Detection in UK Biobank Chromosome 20 Data
False-positive rates for identity by descent (IBD) segment detection for 5,000, 50,000, and 485,346 UK Biobank samples. IBD segments with length ≥2 centiMorgan (cM) were detected with each method. Detected IBD segments were assigned into bins of 2–3, 3–4, 4–6, 6–10, 10–18, and >18 cM according to their segment length. The false-positive rate is the proportion of detected IBD segments in a bin that are not covered by any true IBD segment >1.5 cM in length. Hap-IBD is the only method shown for the full UK Biobank analysis (485,346 individuals) because other methods were unable to complete the analysis with a 2 cM output threshold within the memory and time constraints (see the Computational Feasibility subsection in Results). The x-coordinate of each data point is the left bin end point (e.g., 2 cM for the 2–3 cM bin). For the full range of y-coordinate values, see Figure S2.