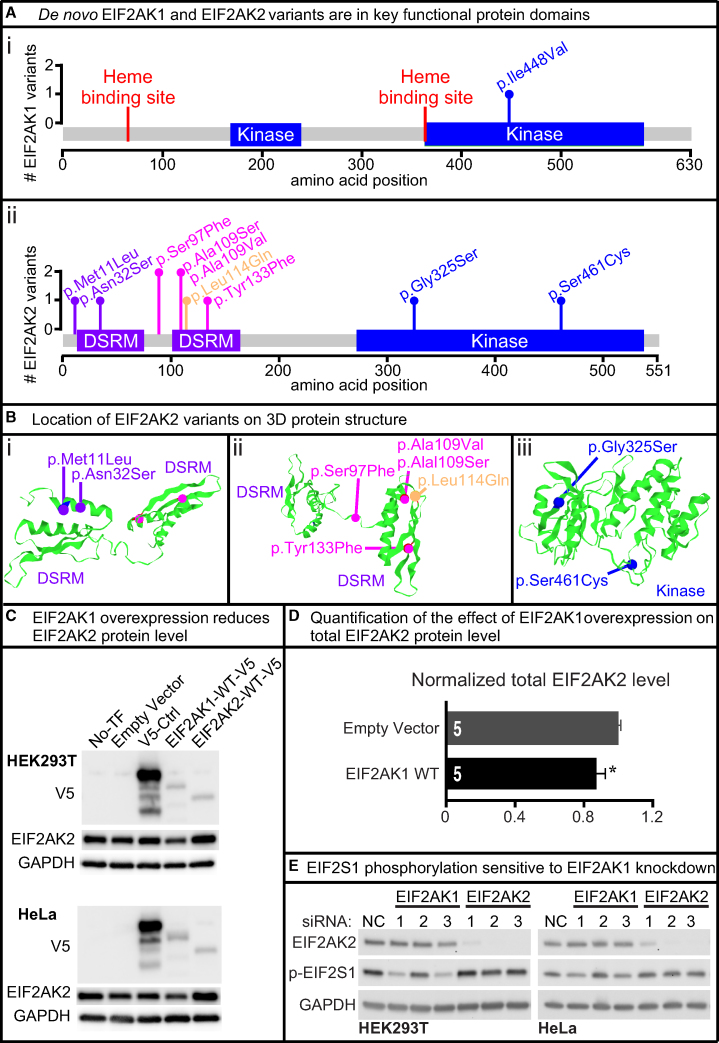
Figure 2.
De novo EIF2AK1 and EIF2AK2 Missense Variants Map to Key Protein Domains and EIF2AK1 Knockdown Impairs EIF2S1 Phosphorylation
(A) Lollipop plots showing variants relative to a schematic representation of the gene adapted from MutationMapper. Heme-binding sites in red, protein kinase domain (Kinase) in blue, and double-stranded RNA-binding motif (DSRM) in purple. EIF2AK1 variant (i) is located in the kinase domain and EIF2AK2 variants (ii) are located in the DSRM and Kinase domains.
(B) 3D structure of EIF2AK2 DSRM and Kinase domains with de novo EIF2AK2 variants in purple, magenta, or blue. The rare variant control, p.Leu114Gln, is in orange. Variants are mapped to the protein 3D structure using Mutation3D.49 PDB: 1QU6, 3UIU.
(C) Full-length human EIF2AK1, EIF2AK2, and unrelated control cDNAs were cloned into pcDNA-DEST40 Vector with a CMV promoter and C terminus V5 tag. Lipofectamine 3000 was used to transfect the cDNA vectors into HEK293T and HeLa cells. Western blots show the protein level of V5-tagged and endogenous EIF2AK2. Increased EIF2AK1 protein level reduces EIF2AK2 protein level in HEK293T and HeLa cell lines. All western blot images in this paper were acquired using the Bio-Rad ChemiDoc Imaging Systems and densitometric analyses of the bands were performed with ImageJ. All images were collected by the imaging system within the linear range.
(D) Quantification of the effect of increased EIF2AK1 in mammalian cell lines on EIF2AK2 protein levels. Statistical significance determined by Student’s t test. Data shown as mean ± SEM; n = 5 independent replicates. ∗p < 0.05.
(E) Lipofectamine RNAiMAX was used to transfect HEK293T and HeLa cells with either control, EIF2AK1, or EIF2AK2 siRNA for 3 days. Three different siRNAs were tested per gene. Western blots show knockdown efficiency for EIF2AK1 and EIF2AK2. Two EIF2AK1 siRNAs show reduced p-EIF2S1 levels in both HEK293T and HeLa cells. Knockdown of EIF2AK2 does not affect p-EIF2S1 levels in either HEK293T or HeLa cells.