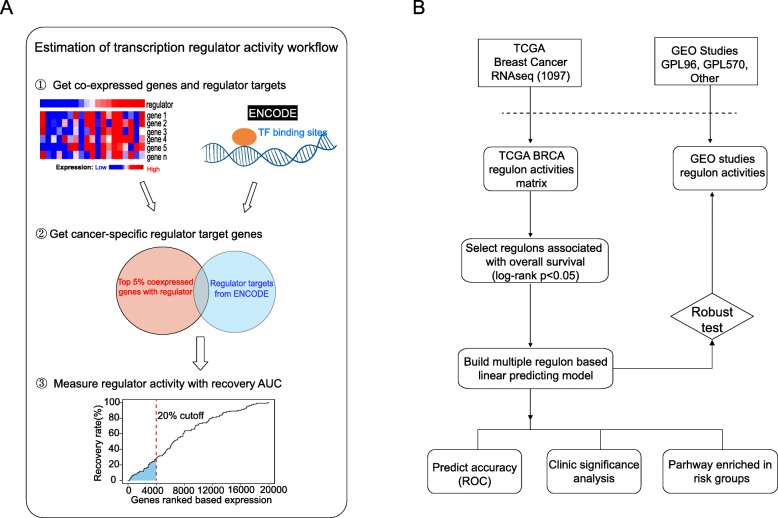
Fig. 1.
Workflow of the study. a. Estimation of transcription regulator activities workflow. Co-expression analysis of each regulator with all genes in TCGA breast cancer cohort was performed using GRNBoost2 and regulator binding site information was retrieved from ENCODE (step 1). The intersection of the top 5% of co-expressed genes with regulator target genes was identified (step 2) and used to calculate the regulator activity with a rank-based approach (step 3). b. Overall workflow of the study. Abbreviations: TCGA, The Cancer Genome Atlas; GEO, Gene Expression Omnibus