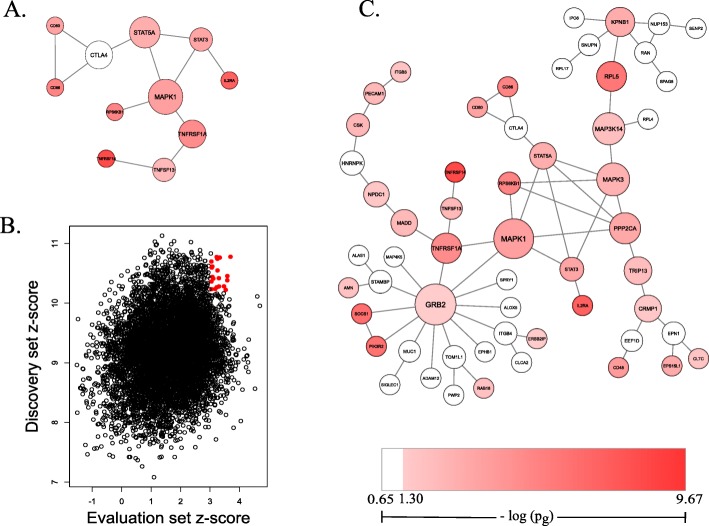
Fig. 3.
Significant modules of dual evaluation regarding IMSGC GWAS as discovery set. The top module of dual evaluation shows MAPK1 as central (a). Modules are regarded as significant based on normalized evaluation and discovery z-scores. Significant modules are represented in red (b). The 20 significant modules were merged to show overall gene network (n = 56) (c). Large nodes represent central genes and small nodes represent less central genes (based on measure of Betweenness). Colors of nodes represent Pascal reported gene-level p-values as specified by legend