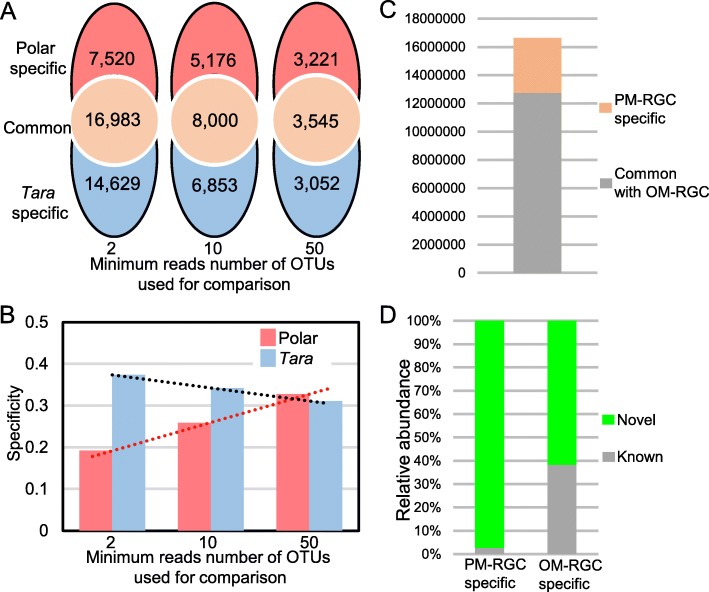
Fig. 4.
Comparison between the polar microbiomes and the Tara Oceans data. a Venn diagram showing the distribution of operational taxonomic units (OTUs) across the polar and the Tara miTags. The Tara miTags comprise 16S rRNA sequences extracted from 243 seawater metagenomes. Venn diagrams based on OTUs with more than 2, 10, 50 miTag numbers are shown. b The change of the taxonomic specificity along with the OTU abundances (indicated by the reads number of OTUs) included for comparison. c Establishment of a polar microbial reference gene catalog (PM-RGC) and comparison with the ocean microbial reference gene catalog (OM-RGC) established by the Tara Oceans study. PM-RGC specific gene orthologs are nonredundant genes that are present in the polar seawater metagenomes but were not detected in the OM-RGC. d BLASTp searching the NCBI-Nr database using the PM-RGC or OM-RGC specific gene catalog. Genes got hit in NCBI-Nr database are labeled as “known” while genes with no hit are labeled as “novel”