Fig. 2.
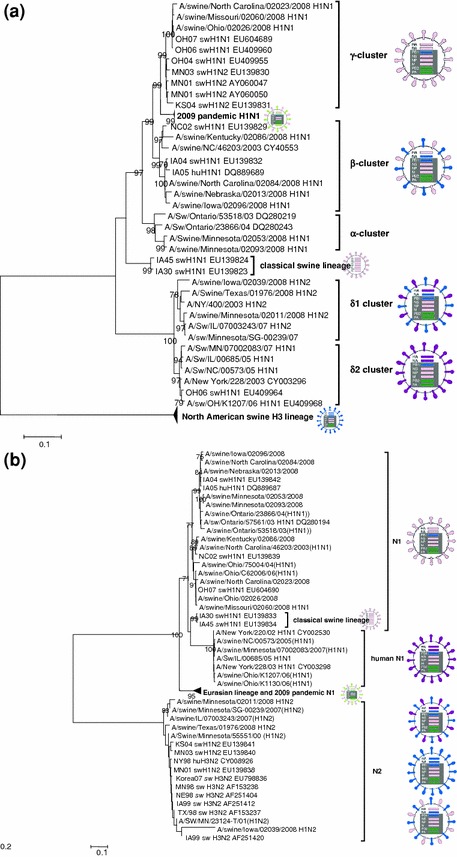
Neighbor-joining trees inferred from multiple nucleotide sequence alignment of segment 4 (HA, a) and segment 6 (NA, 2b). a shows four H1 clusters of viruses, H1α, H1β, H1γ and H1δ (human-like H1) as indicated by the bars on the right of the tree. In both trees, the HA cluster specificity is indicated. The genomic constellation of each clade is indicated by the images on the right side of the tree. Classical swine lineage is color coded pink, avian lineage is coded green, human lineage is coded blue or purple. Light green indicates the Eurasian avian/swine lineage. Classical swine lineage-HA gene (a) was acquired by the TRIG cassette and evolved overtime to form α-, β- and γ- clusters. The introduction of human seasonal HA from H1N2 and H1N1 gave rise to δ cluster viruses differentiated phylogenetically by two distinct sub-clusters, δ1 and δ2 (a). Similar to the δ-cluster viruses in the HA phylogenetic analysis, β-viruses have split into two sub-clusters (b). Phylogenetic analyzes were conducted in MEGA4. Statistical support was provided by bootstrapping over 1,000 replicates and bootstrap values >70 are indicated at the correspondent node. The scale bars indicate the estimated numbers of nucleotide substitutions per site. human (Hu), swine (Sw)
