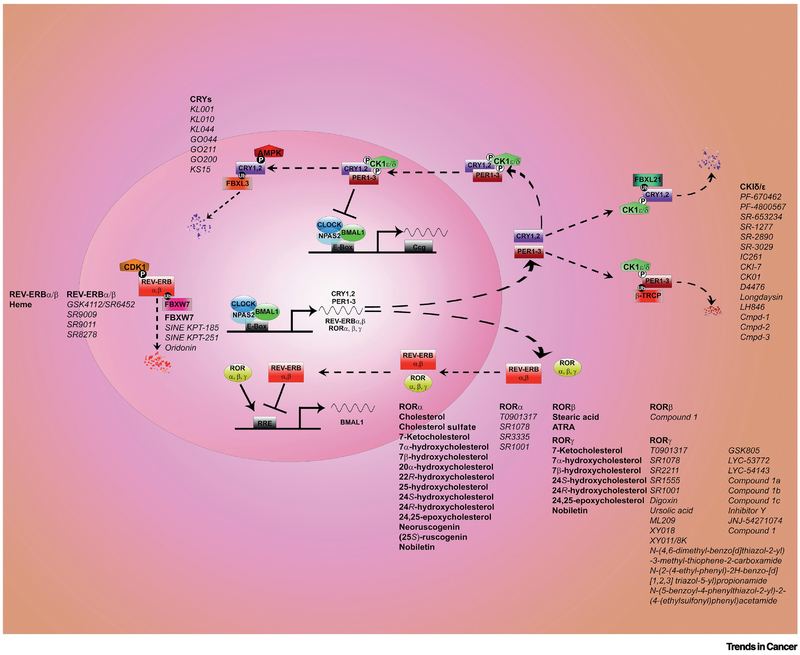
FIGURE 1. Circadian clock: molecular mechanisms, natural ligands and, chemical tools.
The circadian clock machinery generates everyday circadian rhythms. The core clock mechanism presents activators and repressors and consists of two main loops. Several regulatory layers (transcriptional, post-translational, cytoplasm-nuclear translocation) are in play to ensure rhythmicity. Transcriptional activators BMAL1 and CLOCK/NPAS2 (NPAS2 substitutes for CLOCK in the forebrain) bind to DNA element E-box motif and trigger the expression of the repressors Period (PER1, PER2, PER3), Cryptochrome (CRY1 and CRY2). Upon accumulation PERs and CRYs interact, they are phosphorylated by Casein Kinase 1δ/ε and translocate to the nucleus where they block the transcriptional activity of BMAL1 and CLOCK/NPAS2 thus inhibiting their own transcription. Post-translational mechanisms driven by AMPK and FBXL3 will lead CRYs to degradation in the nucleus; cytoplasmic degradation of CRYs is instead regulated by CK1δ/ε and FBXL21. Several chemical tools have been developed to modulate CRYs and CK1δ/ε as displayed. PERs are degraded in the cytoplasm upon phosphorylation by CK1δ/ε and β-TRCP. In an additional loop BMAL1 and CLOCK/NPAS2 activate the transcription of REV-ERBα/β and RORα/β/γ proteins. These are respectively transcriptional repressors and activators that further ensure rhythmicity of BMAL expression. Both REV-ERBα/β and RORα/β/γ are nuclear hormone receptors that are modulated by natural ligands (in bold in the figure). REV-ERBα/β and RORα/β/γ are rapidly becoming a target of interest for pharmacology and several small molecules (in italic in the figure) have been developed in the recent years to modulate their activity.