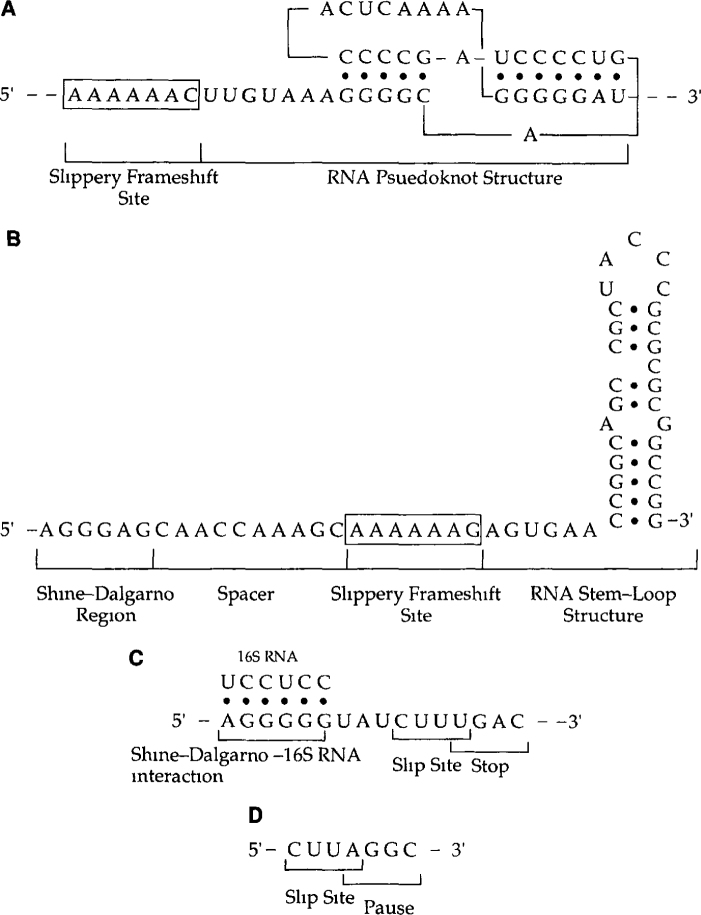
Fig. 2 The signals that cause frameshifting in four different systems. (A) MMTV gag-pro frameshift signal −1 simultaneous slippage frameshifting in mouse mammary tumour virus (MMTV). (B) dnaX frameshift signal −1 dual tRNA slippage in the E. coli dnaX gene. (C) prf frameshift signal +1 frameshifting in the release factors gene prfB in E. coli. (D) Ty1 frameshift signal. +1 frameshifting in retrotransposon Ty1 in yeast Saccharomyces cerevisiae All the frameshift events shown require a pause that is provided by a pseudoknot (A), stem-loop structure (B), stop codon (C), or a slowly decoded codon (D) as well as require a slippery sequence. Frameshifting in the dnaX (B) and the prfB (C) genes also require interaction between the Shine-Dalgarno sequence and the complementary sequence in the 16s rRNA.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.