Fig. 15.6.
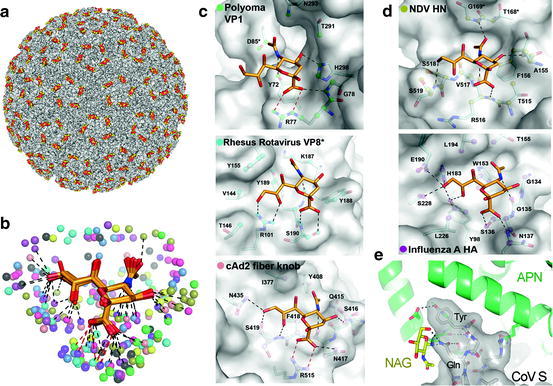
Carbohydrates as viral receptors. (a) Multimeric binding of polyomavirus to its sialic acid receptor. Surface representation of the crystal structure (PDB ID 1SIE) of a polyomavirus particle (surface in grey) in complex with a sialic acid receptor fragment (carbons in yellow). (b) Possible contacts of viral proteins with terminal sialic acid, based on crystal structures [62]. Sialic acid is shown as sticks with carbons in orange. Viral atoms within 4.0 Å of the carbohydrate receptors in crystal structures of virus-sialic acid complexes are shown as spheres. Hydrogen bonds and salt bridges are indicated by black and red lines, respectively. (c) Non-enveloped viruses binding to sialic acid receptors. Detail of sialic acid binding interactions with polyoma VP1 (PDB ID 1VPS), rhesus rotavirus VP8* (PDB ID 1KQR) and cAd2 fibre knob (PDB ID 2WBV). (d) Enveloped viruses binding to sialic acid receptors. Network of interactions for the NDV-HN (PDB ID 1USR) and influenza HA protein (PDB ID 1HGG). (e) Coronavirus (CoV) binding to a glycan-containing region of its aminopeptidase N (APN) receptor. Detail of the interaction revealed by the crystal structure (PDB ID 4F5C) of a fragment of a porcine CoV spike protein (CoV S) in complex with the pig APN. CoV S is shown as grey surface and stick representations, the APN as green ribbons, with the key virus-binding N-acetyl-glucosamine (NAG) residue as sticks and carbons in yellow. The viral Tyr and Gln residues that contact the glycan in the CoV S protein are labelled. Hydrogen bonds are dashed black lines. Oxygens are red and nitrogens dark blue in all figures (Figures (b-d) were provided by U. Neu and T. Stehle, adapted with permission from [62])
