Fig. 2.
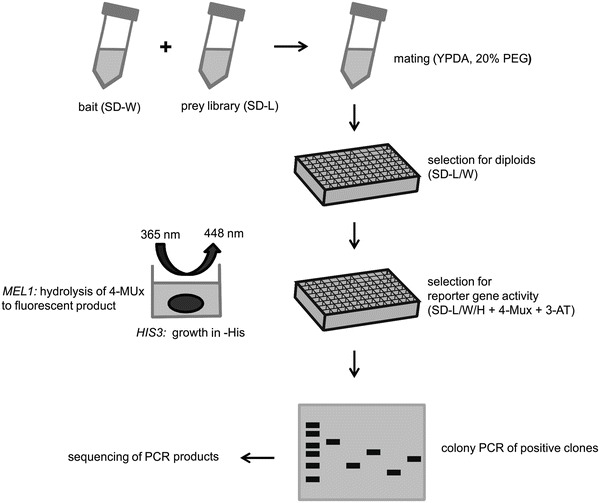
The library-based yeast two-hybrid (Y2H) screening is applied to test an individual bait protein for interaction against a complex prey library, usually consisting of cDNA of a certain tissue type and pretransformed in yeasts. Haploid yeast strains of opposing mating type carrying either bait or prey vectors are propagated under selective conditions in synthetic minimal medium lacking Tryptophane (SD-W) or Leucine (SD-L), respectively. The mating process to gain diploid cells containing both bait and prey vector is performed in complete medium (YPDA + 20 % Polyethyenglycol) under gentle shaking. Afterwards, the yeast culture is spread on 96-well microtiter plates (MTP) and incubated under selective conditions (SD-L/W). The reporter gene activity is then examined in SD medium lacking additionally Histidine (SD-L/W/H) and supplemented with 4-Methylumbelliferyl-α-d-galactopyranoside (4-MUx) and 3-Amino-1,2,4-triazole (3-AT ) in a fluorescence plate reader (excitation 365 nm, emission 448 nm). The interaction of the bait protein fused to the Gal4 DNA binding domain with a prey protein fused to the Gal4 activation domain leads to reconstitution of the Gal4 transcription factor and subsequently expression of HIS3 and MEL1 reporter genes. The reporter genes allow growth in a Histidine-free environment as well as the hydrolysis of 4-MUx to fluorescent Methylumbelliferone, whose fluorescent activity is a measure of protein-protein interaction. The optimal 3-AT concentration as a competitive inhibitor to leaky reporter gene HIS3 expression was defined in a previously performed prescreen. To identify the prey genes that resulted in a positive outcome, a colony PCR of the respective diploid cells is carried out. The agarose gel electrophoresis reveals those hits, where only one prey gene is present. These PCR products then undergo sequencing analysis and their gene ID is identified using an alignment tool like NCBI’s BLAST (http://blast.ncbi.nlm.nih.gov/)
