Fig. 2.
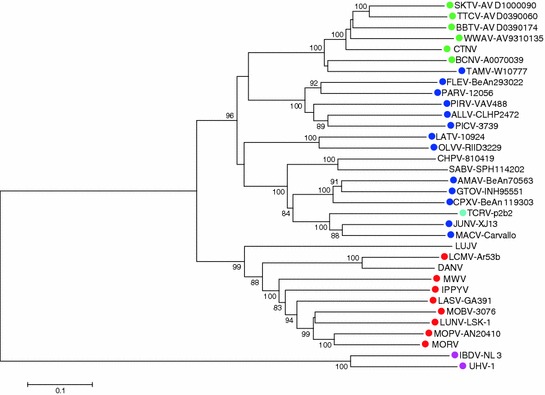
Rooted tree of arenavirus S segment. Phylogeny reconstruction by neighbor joining with 10,000 bootstrap replications under maximum composite likelihood substitution model was done with Mega 6 (Tamura et al. 2013). Bootstrap values below 80 % are hidden. A similar tree was obtained for the L segment. GenBank accession numbers of S segment (all complete sequences) used to generate the tree, as well as species names of rodent reservoirs were obtained from previously published data (King et al. 2012), with the following exceptions: Skinner tank virus (SKTV, EU123328.1) from Neotoma mexicana (USA); Tonto creek virus (TTCV, EF619034.1) from Neotoma albigula; Big brushy tank virus (BBTV, EF619035.1) from Neotoma albigula, Arizona (USA); Catarina virus (CTNV, DQ865244) from Neotoma micropus (Cajimat et al. 2013); and Luna virus (LUNV-LSK-1, AB693148.1) from Mastomys natalensis, Zambia (Ishii et al. 2012). Colored dots next virus isolates indicate reservoir species: green, neotominae subfamily; blue, sigmodontinae subfamily; red, murinae subfamily; cian, the bat Artibeus spp, proposed reservoir of Tacaribe virus (TCRV-p2b2); and magenta, the Boa constrictor snake, reservoir of the recently reported putative arenaviruses University of Helsinki virus (UHV-1) and inclusion body disease virus (IBDV-NL3) (Bodewes et al. 2013; Hetzel et al. 2013). Absence of colored dot indicates unknown reservoir
