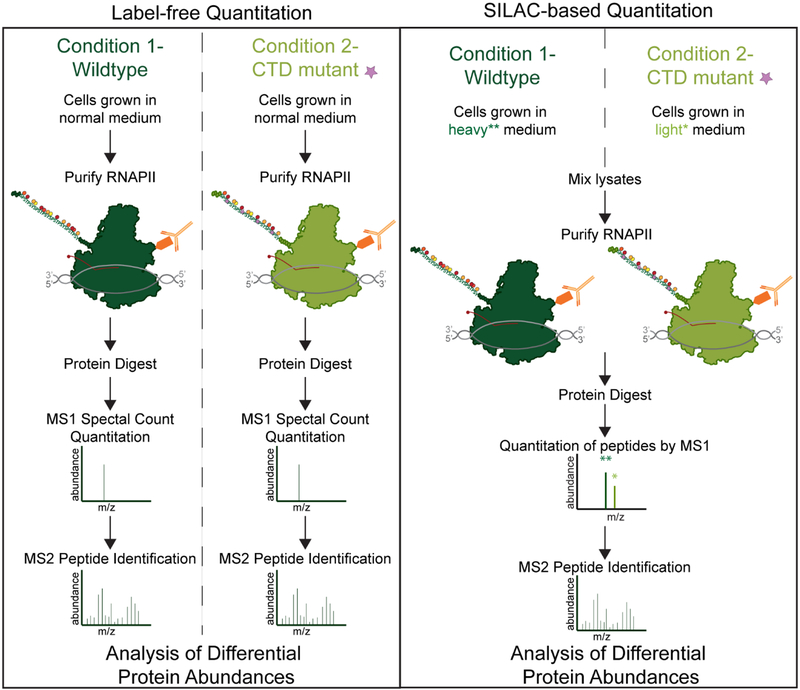
Figure 3: Strategies for Quantitative Analysis of RNAPII interactors.
A) Label-free quantitation of RNAPII associated proteins can be done from multiple conditions. Individual mass spectrometry is performed and then quantitation is performed by either MS1 precursor quantitation of ion intensity and/or MS2 fragment ion count based quantitation (referred to as spectral counting or peptide-spectrum match (PSM) counting). B) SILAC based quantitation can be performed using cells grown in various combinations of light and heavy amino acids typically focused on lysine and arginine. Samples can be mixed together based on equal cell numbers or by equal protein concentration. Quantitation is then performed by analysis of the heavy isotope containing or light isotope containing MS1 ion intensities. MS2 based fragment ion analysis is still used for peptide fingerprinting but is not used for quantitation in SILAC based approaches.