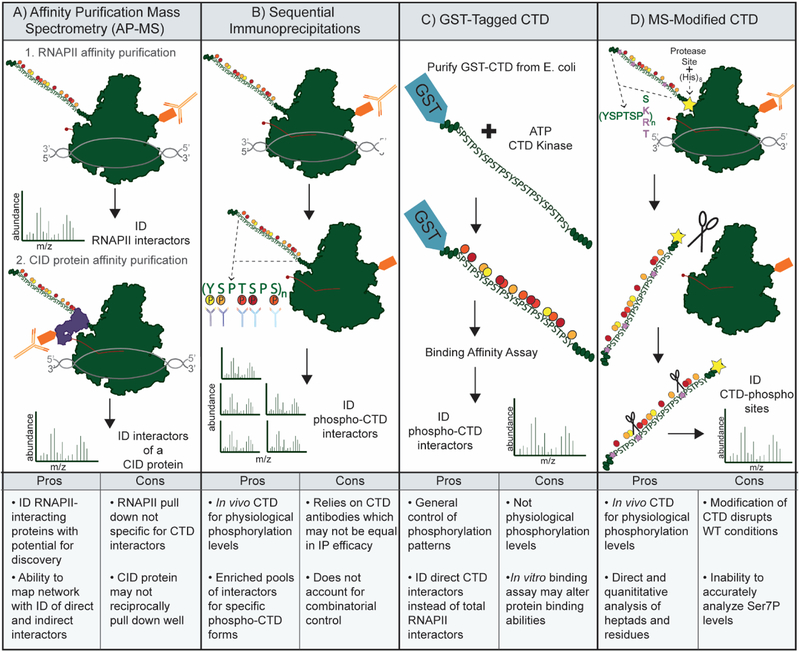
Figure 4: Strategies for Analysis of the CTD and CTD interactors.
A) Affinity Purification Mass Spectrometry (AP-MS) of RNAPII complexes allows identification (ID) of RNAPII interactors. Interactors of interest with CTD Interaction Domains (CIDs) may then be purified to analyze the CTD interactome. B) Purification of total RNAPII followed by IPs using antibodies specific for CTD phospho-sites. MS analysis to identify protein interactome for CTD phospho-forms. C) Purification of GST-tagged CTD peptides to be phosphorylated by CTD kinases and used in binding assays. MS analysis of bound proteins to ID CTD-interacting proteins. D) Modification of the CTD in cells to increase suitability for MS analysis. Mutation of select Ser7 residues to lysine to provide trypsin cleavage sites, as well as additional mutations to create unique mass peptides. MS analysis of these msCTDs allows mapping of specific post-translational modifications.