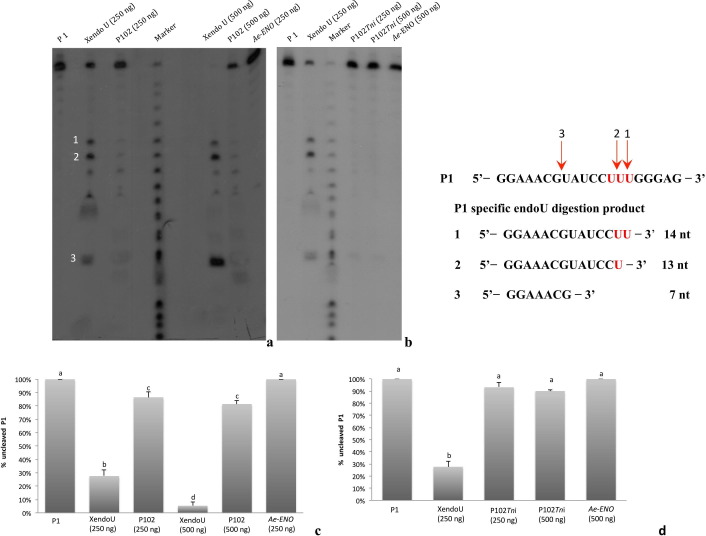
Fig. 5.
Analysis of the endoribonuclease-U activity of P102 (a) and P102Tni (b). Assay performed with 250 ng or 500 ng of each protein (P102 and P102Tni) with 5′-32P-labeled oligoribonucleotide P1, displayed only a slight residual activity. The greater quantity of enzymes (500 ng) did not show any significant differences in the catalytic activity of P102 and P102Tni. Labeled oligoribonucleotide P1 incubated in absence of enzymes was loaded in the first lanes (a, b). The XendoU enzyme (250 ng and 500 ng) and an unrelated protein, the Ae-ENO from Aphidius ervi (250 ng and 500 ng) were used as positive and negative controls, respectively. Marker: nucleotide ladder, generated by alkaline digestion of P1 oligoribonucleotide (Laneve et al., 2003). Densitometric analysis of P102 (c), P102Tni (d) and controls: enzymatic activity is expressed as relative surface area corresponding to band intensity of the uncleaved RNA (oligoribonucleotide P1 substrate) in each lane. Error bars represent the standard error of the mean for three independent experiments. Significant differences are denoted by different letters (Tukey’s test, p < 0.05).