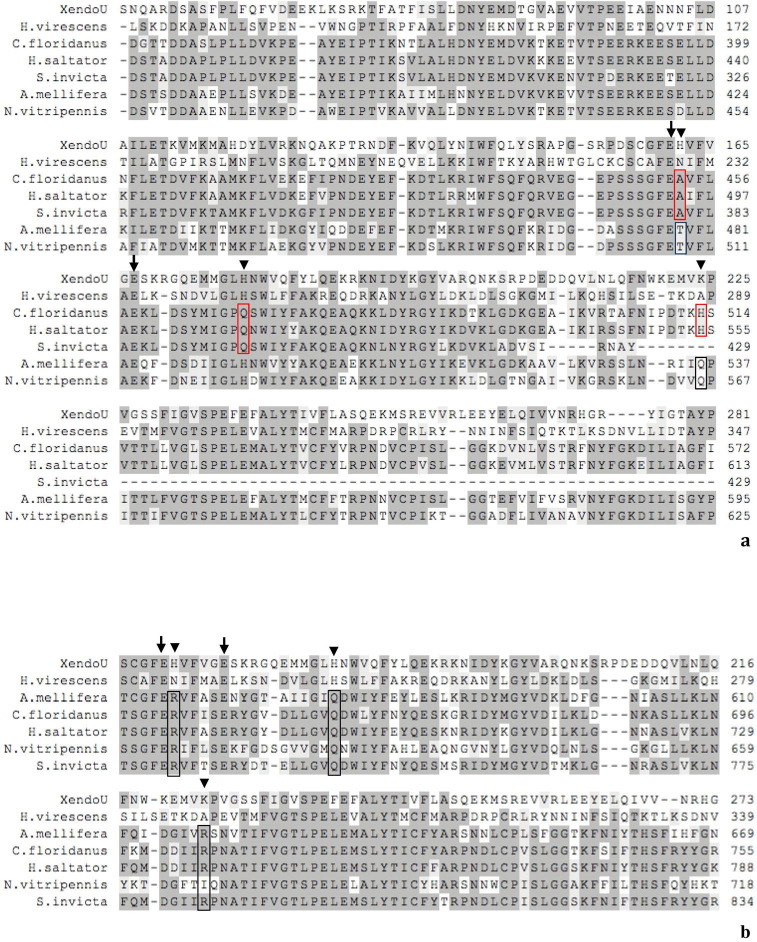
Supplementary Fig. 6.
Partial alignment of XendoU (residues 48-281 (a) residues 157-273 (b)) and P102 sequences with homologs from Hymenoptera (identity with P102 about 40%) not containing the canonical XendoU residues in the catalytic site. Three different classes of variants of the five catalytic residues compared to XendoU are shown. Simultaneous substitution of the histidines at positions H162 and H178 in alanine and glutamine respectively and the lisyne at position K224 in histidine (red box), discovered only in Hymenoptera Vespoidea (Camponotus floridanus, GenBank ID: EFN67408.1; Harpegnathos saltator, GenBank ID: EFN88107.1; Solenopsis invicta, GenBank ID: EFZ20078.1) and substitution of the histidine at position H162 in threonine and of lysine at position K224 in glutamine (blue box) in Hymenoptera Apoidea and Chalcidoidea (Apis mellifera, RefSeq ID: XP_625112.2; Nasonia vitripennis, RefSeq ID: XP_001606738.1)(a). In all three families the histidines were identified at positions H162 and H178 and of lysine at position K224 varied in arginine, glutamine and arginine or isoleucine, respectively (black box) (A. mellifera, RefSeq ID: XP_625106.3; C. floridanus, GenBank ID: EFN67407.1; H. saltator, GenBank ID: EFN88106.1; N. vitripennis, RefSeq ID: XP_003424318.1; S. invicta, GenBank ID: EFZ20057.1) (b). Residues involved in the enzymatic activity are highlighted: arrows mark the conserved residues of the catalytic site while arrowheads indicate the alternative ones.