Fig. 6.
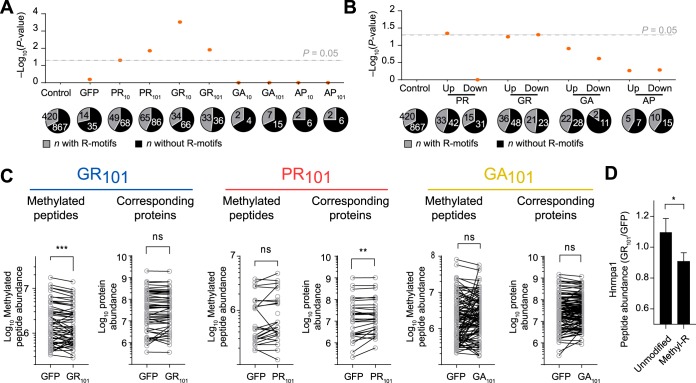
Proteome hypomethylation upon GFP-GR101 expression. A, Shown are p values of Fisher exact tests for enrichment of arginine motifs (34) in the DPR interactomes versus Control, which are the background proteins observed in the whole proteome analysis and which were not deemed significantly affected in abundance by DPR expression. Numbers of proteins are indicated. B, As for A, but for proteins seen significantly up-regulated and downregulated in total protein lysate compared with Control. C, Abundances of methylated peptides seen in the whole proteome, and the corresponding protein abundances from which they derive. The data is plotted as matched-pairs of peptides (or proteins) with differences evaluated by 2-tailed Wilcoxon signed-rank test. p values are coded as ns > 0.05; **, p < 0.01; ***, p < 0.001. The mean difference in abundances of matched pairs of methylated peptides (GR101 - GFP) is −399,018. D, Shown are the means ± S.E. of peptide abundance ratios of Hnrnpa1 from the PR101 and GFP samples. Shown are unmodified (n = 24) and arg-methylated peptides (n = 5). Significance of difference was assessed with an unpaired t test with Welch's correction. p value is coded as *, p < 0.05.