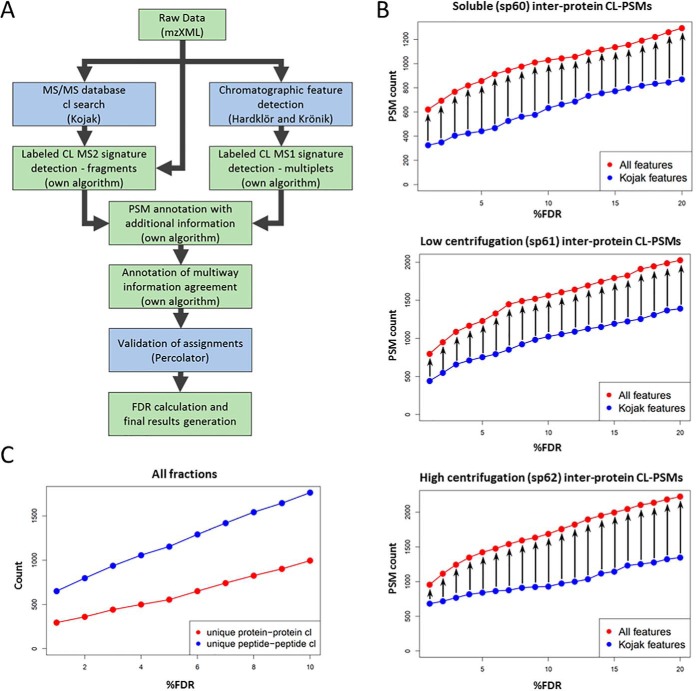
Fig. 4.
Crosslinker-specific mass spectrum features improve crosslinker-modified peptide identification. A, MS data is passed into our software pipeline that generates PSMs, extracts MS1 feature information, adds additional CL-specific feature information to each PSM, executes PSM validation, and returns validated PSMs. B, The number of identified inter-protein CL-PSMs as a function of %FDR are shown for PSM validation outputs from Percolator training with input using either the original set of Kojak PSM validation features, or the full set of PSM validation features from the data analysis pipeline. An increase in identified CL-PSMs was observed across all %FDR levels (0–20% shown). C, The total number of unique protein-protein interactions and unique residue-residue crosslink identifications in all datasets combined is shown as a function of %FDR.