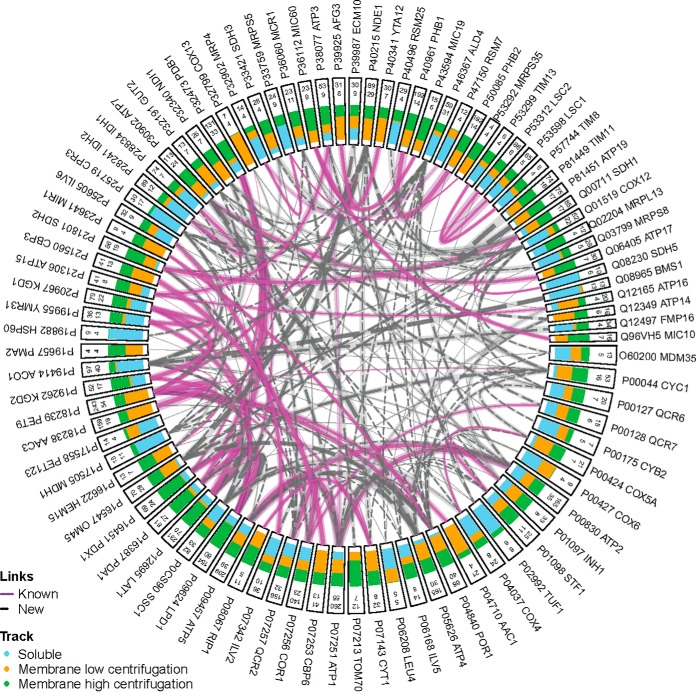
Fig. 6.
Protein-protein interaction network analysis and sub-compartment localization of the identified crosslinks. Unique inter-protein crosslinks identified at 2% FDR are represented for those proteins with a minimum of 4 unique residue-residue crosslinks. Classification of a protein interaction (edges) as “known” or “new” was based on the EMBL-EBI IntAct database of known yeast mitochondrial protein-interactions (retrieved: October 18, 2019) (45). Starting from the outside, each node is labeled with the UniProtKB accession number followed by the gene name. The number on the outside represents the total number of PSMs associated with that protein, followed by the number of unique residue-residue crosslinks associated with that protein. The green, orange, and blue bars inside each rectangle indicate sample pre-fractions in which the respective protein was identified. The width of the edges (i.e. the lines connecting nodes) represents the proportional number of all PSMs associated with the respective nodes with the number of validated PSMs between two proteins represented in semi-transparent highlighting and the number of unique residue-residue pairs represented by solid lines.